6DJ3
 
 | |
3G35
 
 | |
3D6A
 
 | | Crystal structure of the 2H-phosphatase domain of Sts-2 in complex with tungstate. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Sts-2 protein, ... | | Authors: | Chen, Y, Carpino, N, Nassar, N. | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of the 2H-phosphatase domain of Sts-2 reveals an acid-dependent phosphatase activity.
Biochemistry, 48, 2009
|
|
3G32
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 6 (3G3) | | Descriptor: | 2-[2-(1H-tetrazol-5-yl)ethyl]-1H-isoindole-1,3(2H)-dione, Beta-lactamase CTX-M-9a, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, Y, Shoichet, B.K. | | Deposit date: | 2009-02-01 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Molecular docking and ligand specificity in fragment-based inhibitor discovery
Nat.Chem.Biol., 5, 2009
|
|
3G2Y
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 1 (GF4) | | Descriptor: | 4-ethyl-5-methyl-2-(1H-tetrazol-5-yl)-1,2-dihydro-3H-pyrazol-3-one, Beta-lactamase CTX-M-9a, DIMETHYL SULFOXIDE | | Authors: | Chen, Y, Shoichet, B.K. | | Deposit date: | 2009-02-01 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Molecular docking and ligand specificity in fragment-based inhibitor discovery
Nat.Chem.Biol., 5, 2009
|
|
3FKV
 
 | | AmpC K67R mutant complexed with benzo(b)thiophene-2-boronic acid (bzb) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Chen, Y, McReynolds, A, Shoichet, B.K. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Re-examining the role of Lys67 in class C beta-lactamase catalysis.
Protein Sci., 18, 2009
|
|
3FN3
 
 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
3G34
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 11 (1CE) | | Descriptor: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Beta-lactamase CTX-M-9a, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, Y, Shoichet, B.K. | | Deposit date: | 2009-02-01 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Molecular docking and ligand specificity in fragment-based inhibitor discovery
Nat.Chem.Biol., 5, 2009
|
|
6B5H
 
 | |
6B5G
 
 | |
6B5I
 
 | |
6MN6
 
 | |
2IKQ
 
 | |
6N7E
 
 | |
6KI3
 
 | | The crystal structure of AsfvAP:dF commplex | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*GP*CP*AP*GP*CP*GP*TP*CP*C)-3'), DNA (5'-D(P*(3DR)P*CP*GP*AP*CP*GP*AP*G)-3'), ... | | Authors: | Chen, Y, Gan, J. | | Deposit date: | 2019-07-17 | | Release date: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
2FFY
 
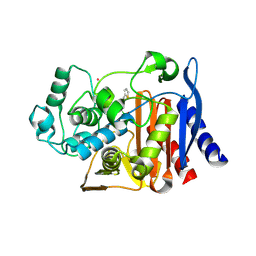 | | AmpC beta-lactamase N289A mutant in complex with a boronic acid deacylation transition state analog compound SM3 | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-PHENYLMETHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Chen, Y, Minasov, G, Roth, T.A, Prati, F, Shoichet, B.K. | | Deposit date: | 2005-12-20 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution
J.Am.Chem.Soc., 128, 2006
|
|
7YA2
 
 | |
7YOV
 
 | |
7YOT
 
 | |
7YOU
 
 | |
5E9T
 
 | | Crystal structure of GtfA/B complex | | Descriptor: | Glycosyltransferase Gtf1, Glycosyltransferase-stabilizing protein Gtf2, MAGNESIUM ION | | Authors: | Chen, Y, Rapoport, T.A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Mechanism of a cytosolic O-glycosyltransferase essential for the synthesis of a bacterial adhesion protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8I4B
 
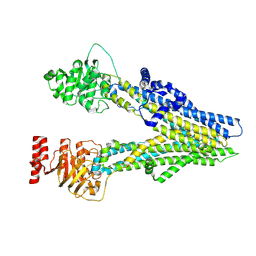 | | Cryo-EM structure of apo-form ABCC4 | | Descriptor: | ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4A
 
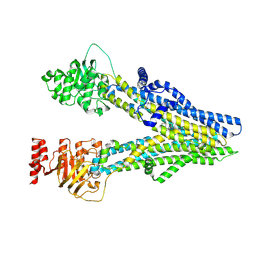 | | Cryo-EM structure of dipyridamole-bound ABCC4 | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4C
 
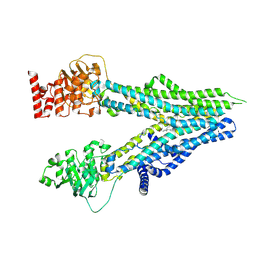 | | Cryo-EM structure of U46619-bound ABCC4 | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8J3Z
 
 | | Cryo-EM structure of ATP-U46619-bound ABCC4 | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, ... | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-04-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
