6M22
 
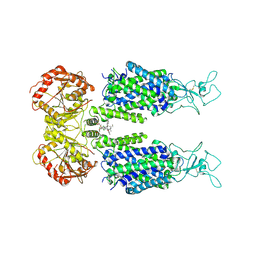 | | KCC3 bound with DIOA | | Descriptor: | 2-[[(2~{R})-2-butyl-6,7-bis(chloranyl)-2-cyclopentyl-1-oxidanylidene-3~{H}-inden-5-yl]oxy]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
7XT6
 
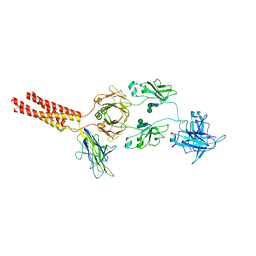 | | Structure of a membrane protein M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, ... | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
6M23
 
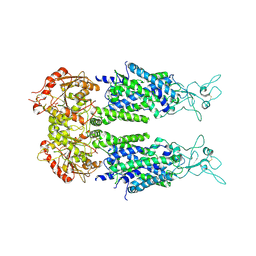 | | Overall structure of KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
6CZ1
 
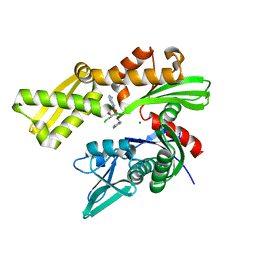 | | Crystal structure of ATPase domain of Human GRP78 bound to Ver155008 | | Descriptor: | 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, Endoplasmic reticulum chaperone BiP, MAGNESIUM ION | | Authors: | Antoshchenko, T, Chen, Y, Hughes, S, Park, H. | | Deposit date: | 2018-04-07 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic selection of adenosine analogs that fit the mold of the active site of human GRP78 and beyond
To be Published
|
|
1XKL
 
 | | Crystal Structure of Salicylic Acid-binding Protein 2 (SABP2) from Nicotiana tabacum, NESG Target AR2241 | | Descriptor: | 2-AMINO-4H-1,3-BENZOXATHIIN-4-OL, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Chen, Y, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6Y8J
 
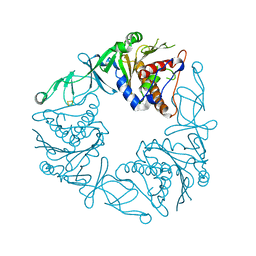 | | Crystal structure of the apo form of a quaternary ammonium Rieske monooxygenase CntA | | Descriptor: | Carnitine monooxygenase oxygenase subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
4RM0
 
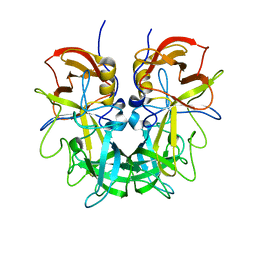 | | Crystal structure of Norovirus OIF P domain in complex with Lewis a trisaccharide | | Descriptor: | Capsid protein, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | Deposit date: | 2014-10-18 | | Release date: | 2015-06-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
4RLZ
 
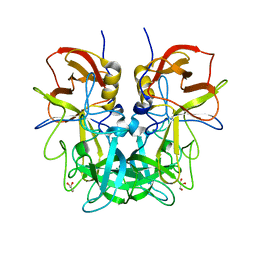 | | Crystal structure of Norovirus OIF P domain | | Descriptor: | Capsid protein, GLYCEROL | | Authors: | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | Deposit date: | 2014-10-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
6Y9D
 
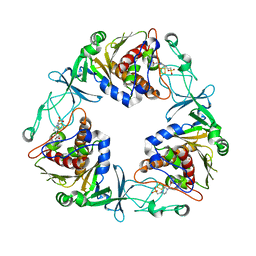 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate L-Carnitine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARNITINE, Carnitine monooxygenase oxygenase subunit, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6Y8S
 
 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate gamma-butyrobetaine | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6YQ7
 
 | | Taka-amylase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
6YQC
 
 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol epoxide inhibitor | | Descriptor: | (1~{R},2~{S},4~{R},5~{S},6~{R})-6-[(2~{S},3~{R},4~{R},5~{S},6~{R})-5-heptoxy-6-(hydroxymethyl)-3,4-bis(oxidanyl)oxan-2-yl]oxy-5-(hydroxymethyl)cyclohexane-1,2,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
6YQ9
 
 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol epoxide inhibitor | | Descriptor: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
6YQB
 
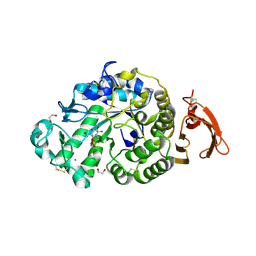 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol cyclosulfate inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
6YQA
 
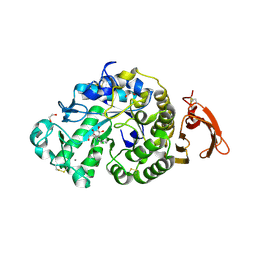 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol aziridine inhibitor | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},5~{R})-5-(8-azanyloctylamino)-4-(hydroxymethyl)cyclohexane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
6Y9C
 
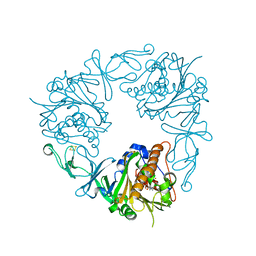 | | The structure of a quaternary ammonium Rieske monooxygenase reveals insights into carnitine oxidation by gut microbiota and inter-subunit electron transfer | | Descriptor: | CARNITINE, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterisation of an unusual cysteine pair in the Rieske carnitine monooxygenase CntA catalytic site.
Febs J., 2023
|
|
4UAI
 
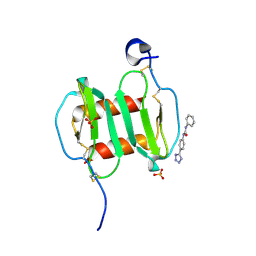 | | Crystal structure of CXCL12 in complex with inhibitor | | Descriptor: | 1-phenyl-3-[4-(1H-tetrazol-5-yl)phenyl]urea, SULFATE ION, Stromal cell-derived factor 1 | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2014-08-09 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Novel Small Molecule Ligand Bound to the CXCL12 Chemokine.
J.Med.Chem., 57, 2014
|
|
6XD7
 
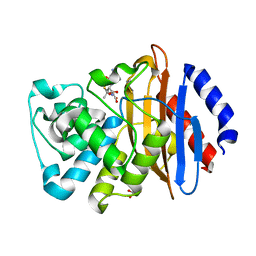 | | KPC-2 N170A mutant bound to hydrolyzed ampicillin at 1.65 A | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | KPC-2 beta-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate.
J.Biol.Chem., 296, 2020
|
|
6XD5
 
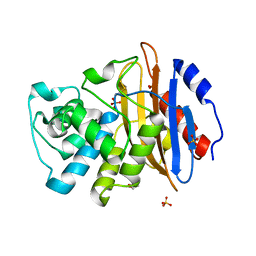 | | Apo KPC-2 N170A mutant at 1.20 A | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, SULFATE ION | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | KPC-2 beta-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate.
J.Biol.Chem., 296, 2020
|
|
6ZGP
 
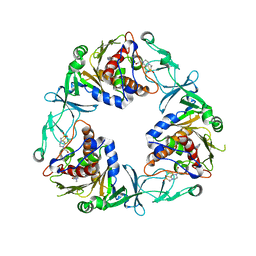 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with inhibitor MMV12 (MMV020670) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D.H, Cameron, A, Chen, Y. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
1VBT
 
 | | Structure of cyclophilin complexed with sulfur-substituted tetrapeptide AAPF | | Descriptor: | CYCLOPHILIN A, SULFUR-SUBSTITUTED TETRAPEPTIDE | | Authors: | Zhao, Y, Chen, Y, Schutkowski, M, Fischer, G, Ke, H. | | Deposit date: | 1998-06-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into Conversion of Substrate to Inhibitor
To be Published
|
|
1M6X
 
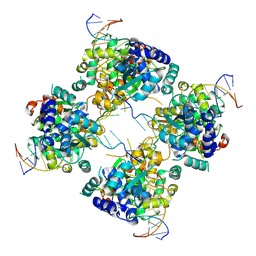 | | Flpe-Holliday Junction Complex | | Descriptor: | Flp recombinase, Symmetrized FRT site | | Authors: | Conway, A.B, Chen, Y, Rice, P.A. | | Deposit date: | 2002-07-17 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Plasticity of the Flp-Holliday Junction Complex
J.Mol.Biol., 326, 2003
|
|
4BBQ
 
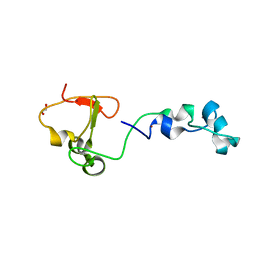 | | Crystal structure of the CXXC and PHD domain of Human Lysine-specific Demethylase 2A (KDM2A)(FBXL11) | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 2A, ZINC ION | | Authors: | Allerston, C.K, Watson, A.A, Edlich, C, Li, B, Chen, Y, Ball, L, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Laue, E.D, Gileadi, O. | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of the Cxxc and Phd Domain of Human Lysine-Specific Demethylase 2A (Kdm2A)(Fbxl11)
To be Published
|
|
6PK0
 
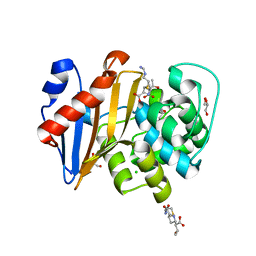 | | Crystal Structure of OXA-48 with Hydrolyzed Imipenem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Substrate Specificity and Carbapenemase Activity of OXA-48 Class D beta-Lactamase.
Acs Infect Dis., 6, 2020
|
|
