216D
 
 | |
6LE6
 
 | | Structure of LNLPTQGRAR bound FEM1C | | Descriptor: | Protein fem-1 homolog C,10-mer peptide, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LBF
 
 | | Crystal structure of FEM1B | | Descriptor: | Protein fem-1 homolog B, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.252 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
4RWT
 
 | | Structure of actin-Lmod complex | | Descriptor: | Actin-5C, Leiomodin-2, MAGNESIUM ION, ... | | Authors: | Chen, X, Ni, F, Wang, Q. | | Deposit date: | 2014-12-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6LDP
 
 | | Structure of CDK5R1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEN
 
 | | Structure of NS11 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,NS11 peptide | | Authors: | Chen, x, Liao, S, Xu, C. | | Deposit date: | 2019-11-25 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LF0
 
 | | Structure of FEM1C | | Descriptor: | Protein fem-1 homolog C, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LBN
 
 | | Structure of SIL1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEY
 
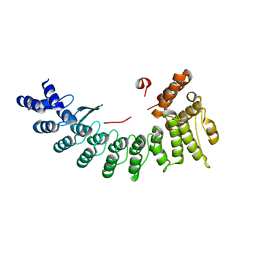 | | Structure of Sil1G bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6Z2F
 
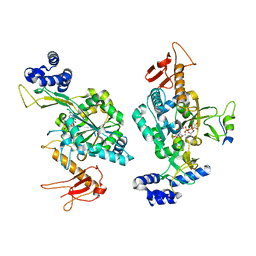 | |
1EGA
 
 | | CRYSTAL STRUCTURE OF A WIDELY CONSERVED GTPASE ERA | | Descriptor: | PROTEIN (GTP-BINDING PROTEIN ERA), SULFATE ION | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1998-12-01 | | Release date: | 1999-07-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3EJJ
 
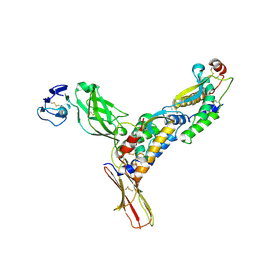 | | Structure of M-CSF bound to the first three domains of FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Colony stimulating factor-1, Macrophage colony-stimulating factor 1 receptor | | Authors: | Chen, X, Liu, H, Focia, P.J, Shim, A, He, X. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4ZHJ
 
 | |
375D
 
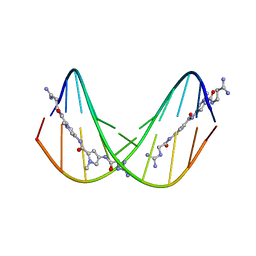 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel End-to-End Binding of Two Netropsins to the DNA Decamers d(CCCCCIIIII) 2, d(CCCBr5CCIIIII)2, d(CBr5CCCCIIIII)2
Nucleic Acids Res., 26, 1998
|
|
2Z59
 
 | | Complex Structures of Mouse Rpn13 (22-130aa) and ubiquitin | | Descriptor: | Protein ADRM1, Ubiquitin | | Authors: | Chen, X, Schreiner, P, Groll, M, Walters, K.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction.
Nature, 453, 2008
|
|
3VPN
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
6XBU
 
 | | polymerase domain of polymerase-theta | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*TP*CP*AP*TP*TP*G)-3'), DNA polymerase theta, ... | | Authors: | Chen, X, Pomerantz, R, Zhao, J. | | Deposit date: | 2020-06-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pol theta reverse transcribes RNA and promotes RNA-templated DNA repair.
Sci Adv, 7, 2021
|
|
3VPM
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
3VPO
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
1KIL
 
 | | Three-dimensional structure of the complexin/SNARE complex | | Descriptor: | Complexin I SNARE-complex binding region, MAGNESIUM ION, SNAP-25 C-terminal SNARE motif, ... | | Authors: | Chen, X, Tomchick, D, Kovrigin, E, Arac, D, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexin/SNARE complex.
Neuron, 33, 2002
|
|
1J51
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM MUTANT (F87W/Y96F/V247L/C334A) WITH 1,3,5-TRICHLOROBENZENE | | Descriptor: | 1,3,5-TRICHLORO-BENZENE, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Chen, X, Christopher, A, Jones, J, Guo, Q, Xu, F, Cao, R, Wong, L.L, Rao, Z. | | Deposit date: | 2002-01-05 | | Release date: | 2002-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the F87W/Y96F/V247L mutant of cytochrome P-450cam with 1,3,5-trichlorobenzene bound and further protein engineering for the oxidation of pentachlorobenzene and hexachlorobenzene
J.BIOL.CHEM., 277, 2002
|
|
1JX6
 
 | | CRYSTAL STRUCTURE OF LUXP FROM VIBRIO HARVEYI COMPLEXED WITH AUTOINDUCER-2 | | Descriptor: | 3A-METHYL-5,6-DIHYDRO-FURO[2,3-D][1,3,2]DIOXABOROLE-2,2,6,6A-TETRAOL, CALCIUM ION, LUXP PROTEIN | | Authors: | Chen, X, Schauder, S, Potier, N, Van Dorsselaer, A, Pelczer, I, BassleR, B.L, Hughson, F.M. | | Deposit date: | 2001-09-05 | | Release date: | 2002-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural identification of a bacterial quorum-sensing signal containing boron.
Nature, 415, 2002
|
|
7K11
 
 | |
7K0Y
 
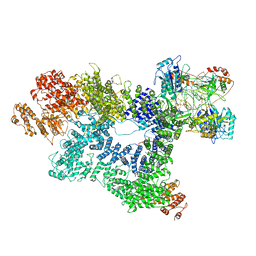 | | Cryo-EM structure of activated-form DNA-PK (complex VI) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-06 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
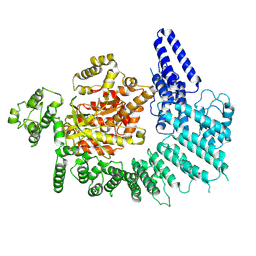
7K10
 
 | |
