8YRP
 
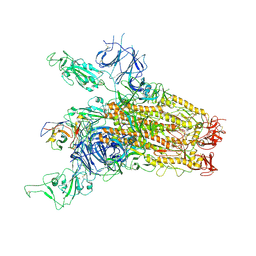 | | SARS-CoV-2 Delta Spike in complex with JM-1A | | Descriptor: | JM-1A Heavy Chain, JM-1A Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YRO
 
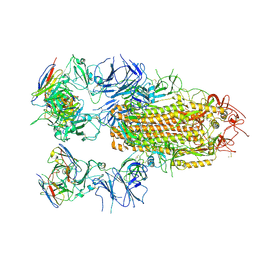 | | SARS-CoV-2 Delta Spike in complex with JL-8C | | Descriptor: | JL-8C Heavy Chain, JL-8C Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
2Q9Q
 
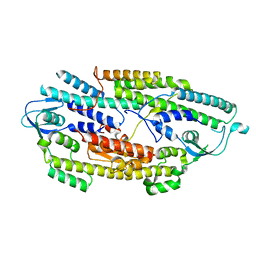 | | The crystal structure of full length human GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, GINS complex subunit 3, ... | | Authors: | Chang, Y.P, Wang, G, Chen, X.S. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of the GINS complex and functional insights into its role in DNA replication.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3BH0
 
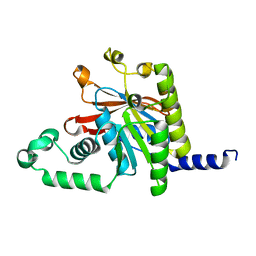 | | ATPase Domain of G40P | | Descriptor: | DNAB-LIKE REPLICATIVE HELICASE | | Authors: | Wang, G, Klein, M.G, Tokonzaba, E, Zhang, Y, Holden, L.G, Chen, X.S. | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of a DnaB-family replicative helicase and its interactions with primase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
7RR5
 
 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
2AOJ
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P6-PR | | Descriptor: | ACETIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
4DYD
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
4E13
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
4E12
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
9ASL
 
 | |
6U8Q
 
 | | CryoEM structure of HIV-1 cleaved synaptic complex (CSC) intasome | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-09-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
9ASK
 
 | |
9ASJ
 
 | |
6KZK
 
 | |
6LQ7
 
 | |
6LQ3
 
 | |
6LPY
 
 | |
6LQ6
 
 | |
6LQ5
 
 | |
6LQ1
 
 | |
6LQ4
 
 | |
6LQ0
 
 | |
6LQ8
 
 | |
