4Q2W
 
 | | Crystal Structure of pneumococcal peptidoglycan hydrolase LytB | | 分子名称: | GLYCEROL, Putative endo-beta-N-acetylglucosaminidase | | 著者 | Bai, X.H, Chen, H.J, Jiang, Y.L, Wen, Z, Cheng, W, Li, Q, Zhang, J.R, Chen, Y, Zhou, C.Z. | | 登録日 | 2014-04-10 | | 公開日 | 2014-07-16 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis.
J.Biol.Chem., 289, 2014
|
|
6IZK
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | 分子名称: | CHLORIDE ION, IMIDAZOLE, L(+)-TARTARIC ACID, ... | | 著者 | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2018-12-19 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus
To Be Published
|
|
5H0S
 
 | | EM Structure of VP1A and VP1B | | 分子名称: | VP1 | | 著者 | Li, X, Zhou, N, Xu, B, Chen, W, Zhu, B, Wang, X, Wang, J, Liu, H, Cheng, L. | | 登録日 | 2016-10-06 | | 公開日 | 2017-01-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
5H0R
 
 | | RNA dependent RNA polymerase ,vp4,dsRNA | | 分子名称: | RNA (42-MER), RNA-dependent RNA polymerase, VP4 protein | | 著者 | Li, X, Zhou, N, Chen, W, Zhu, B, Wang, X, Xu, B, Wang, J, Liu, H, Cheng, L. | | 登録日 | 2016-10-06 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
2ODD
 
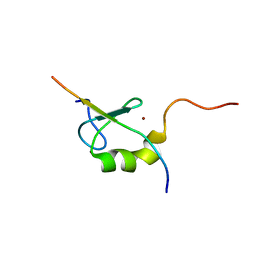 | | Solution structure of the MYND domain from AML1-ETO complexed with SMRT, a corepressor | | 分子名称: | Protein CBFA2T1, SMRT, ZINC ION | | 著者 | Liu, Y.Z, Chen, W, Gaudet, J, Cheney, M.D, Roudaia, L, Cierpicki, T, Klet, R.C, Hartman, K, Laue, T.M, Speck, N.A, Bushweller, J.H. | | 登録日 | 2006-12-22 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity.
Cancer Cell, 11, 2007
|
|
2OD1
 
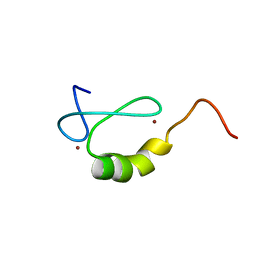 | | Solution structure of the MYND domain from human AML1-ETO | | 分子名称: | Protein CBFA2T1, ZINC ION | | 著者 | Liu, Y.Z, Chen, W, Gaudet, J, Cheney, M.D, Roudaia, L, Cierpicki, T, Klet, R.C, Hartman, K, Laue, T.M, Speck, N.A, Bushweller, J.H. | | 登録日 | 2006-12-21 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity.
Cancer Cell, 11, 2007
|
|
4K6N
 
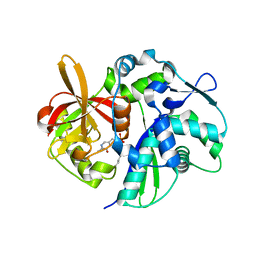 | | Crystal structure of yeast 4-amino-4-deoxychorismate lyase | | 分子名称: | Aminodeoxychorismate lyase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Dai, Y.-N, Chi, C.-B, Zhou, K, Cheng, W, Jiang, Y.-L, Ren, Y.-M, Chen, Y, Zhou, C.-Z. | | 登録日 | 2013-04-16 | | 公開日 | 2013-07-10 | | 最終更新日 | 2013-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and catalytic mechanism of yeast 4-amino-4-deoxychorismate lyase
J.Biol.Chem., 288, 2013
|
|
6UJU
 
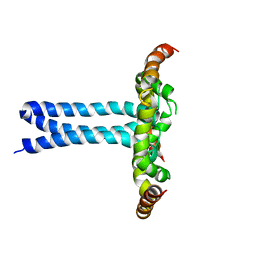 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail (LLP2) | | 分子名称: | Envelope glycoprotein GP41 | | 著者 | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | 登録日 | 2019-10-03 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | 分子名称: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | 著者 | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2017-07-27 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y2W
 
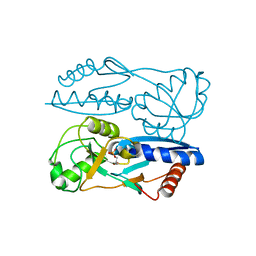 | | Structure of Synechocystis PCC6803 CcmR regulatory domain in complex with 2-PG | | 分子名称: | 2-PHOSPHOGLYCOLIC ACID, Rubisco operon transcriptional regulator | | 著者 | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Cao, D.D, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2017-07-27 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1S9X
 
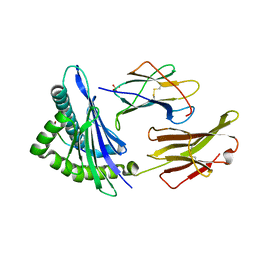 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQA, in complex with HLA-A2 | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | 著者 | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | 登録日 | 2004-02-05 | | 公開日 | 2004-09-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1S9Y
 
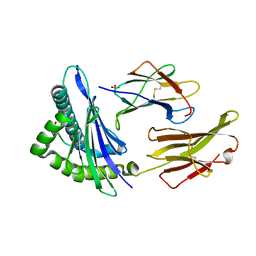 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQS, in complex with HLA-A2 | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | 著者 | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | 登録日 | 2004-02-05 | | 公開日 | 2004-09-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1S9W
 
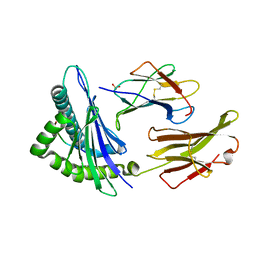 | | Crystal Structure Analysis of NY-ESO-1 epitope, SLLMWITQC, in complex with HLA-A2 | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | 著者 | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | 登録日 | 2004-02-05 | | 公開日 | 2004-09-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1WFC
 
 | | STRUCTURE OF APO, UNPHOSPHORYLATED, P38 MITOGEN ACTIVATED PROTEIN KINASE P38 (P38 MAP KINASE) THE MAMMALIAN HOMOLOGUE OF THE YEAST HOG1 PROTEIN | | 分子名称: | MITOGEN-ACTIVATED PROTEIN KINASE P38 | | 著者 | Wilson, K.P, Fitzgibbon, M.J, Caron, P.R, Griffith, J.P, Chen, W, Mccaffrey, P.G, Chambers, S.P, Su, M.S.-S. | | 登録日 | 1996-09-13 | | 公開日 | 1997-09-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of p38 mitogen-activated protein kinase.
J.Biol.Chem., 271, 1996
|
|
3VTR
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 E328A complexed with TMG-chitotriomycin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | 著者 | Liu, T, Zhou, Y, Chen, L, Chen, W, Liu, L, Shen, X, Yang, Q. | | 登録日 | 2012-06-02 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into cellulolytic and chitinolytic enzymes revealing crucial residues of insect beta-N-acetyl-D-hexosaminidase
Plos One, 7, 2012
|
|
1LV0
 
 | | Crystal structure of the Rab effector guanine nucleotide dissociation inhibitor (GDI) in complex with a geranylgeranyl (GG) peptide | | 分子名称: | GERAN-8-YL GERAN, RAB GDP disossociation inhibitor alpha, SULFATE ION | | 著者 | An, Y, Shao, Y, Alory, C, Matteson, J, Sakisaka, T, Chen, W, Gibbs, R.A, Wilson, I.A, Balch, W.E. | | 登録日 | 2002-05-23 | | 公開日 | 2003-08-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Geranylgeranyl switching regulates GDI-Rab GTPase recycling.
Structure, 11, 2003
|
|
1N67
 
 | | Clumping Factor A from Staphylococcus aureus | | 分子名称: | Clumping Factor, MAGNESIUM ION | | 著者 | Deivanayagam, C.C.S, Wann, E.R, Chen, W, Carson, M, Rajashankar, K.R, Hook, M, Narayana, S.V.L. | | 登録日 | 2002-11-08 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A novel variant of the immunoglobulin fold in surface adhesins of
Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM,
clumping factor A
Embo J., 21, 2002
|
|
6NHW
 
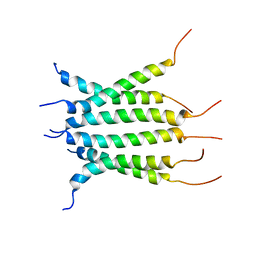 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | 分子名称: | Tumor necrosis factor receptor superfamily member 10B | | 著者 | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | 登録日 | 2018-12-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
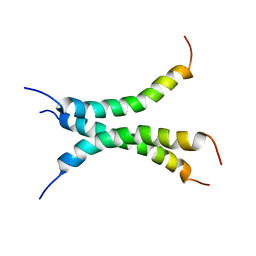 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | 分子名称: | Tumor necrosis factor receptor superfamily member 10B | | 著者 | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | 登録日 | 2018-12-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
2ADU
 
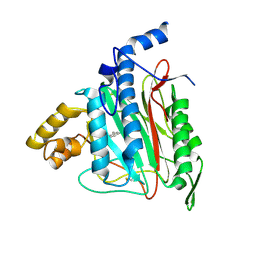 | | Human Methionine Aminopeptidase Complex with 4-Aryl-1,2,3-triazole Inhibitor | | 分子名称: | 4-(3-METHYLPHENYL)-1H-1,2,3-TRIAZOLE, COBALT (II) ION, Methionine aminopeptidase 2 | | 著者 | Kallander, L.S, Lu, Q, Chen, W, Tomaszek, T, Yang, G, Tew, D, Meek, T.D, Hofmann, G.A, Schulz-Pritchard, C.K, Smith, W.W, Janson, C.A, Ryan, M.D, Zhang, G.F, Johanson, K.O, Kirkpatrick, R.B, Ho, T.F, Fisher, P.W, Mattern, M.R, Johnson, R.K, Hansbury, M.J, Winkler, J.D, Ward, K.W, Veber, D.F, Thompson, S.K. | | 登録日 | 2005-07-20 | | 公開日 | 2005-09-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 4-Aryl-1,2,3-triazole: A Novel Template for a Reversible Methionine Aminopeptidase 2 Inhibitor, Optimized To Inhibit Angiogenesis in Vivo
J.Med.Chem., 48, 2005
|
|
4OI3
 
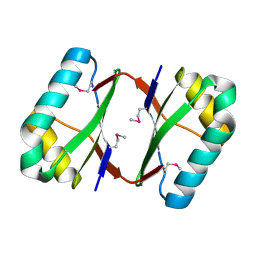 | | Crystal structure analysis of SCO4226 from Streptomyces coelicolor A3(2) | | 分子名称: | Nickel responsive protein | | 著者 | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | 登録日 | 2014-01-18 | | 公開日 | 2014-09-17 | | 最終更新日 | 2014-10-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
4OI6
 
 | | Crystal structure analysis of nickel-bound form SCO4226 from Streptomyces coelicolor A3(2) | | 分子名称: | CITRIC ACID, NICKEL (II) ION, Nickel responsive protein | | 著者 | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | 登録日 | 2014-01-18 | | 公開日 | 2014-09-10 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
8XN9
 
 | | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6 | | 分子名称: | 1D6 VH, 1D6 VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fan, P.F, Ren, Y, Yu, C.M, Chen, W. | | 登録日 | 2023-12-29 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (1.99 Å) | | 主引用文献 | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6
To Be Published
|
|
8XNH
 
 | | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 5C8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5C8-VH, 5C8-VL, ... | | 著者 | Fan, P.F, Ren, Y, Yu, C.M, Chen, W. | | 登録日 | 2023-12-30 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6
To Be Published
|
|
8XC4
 
 | |
