6IZK
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, IMIDAZOLE, L(+)-TARTARIC ACID, ... | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2018-12-19 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus
To Be Published
|
|
6ACL
 
 | |
6ACP
 
 | |
6HEG
 
 | | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpB, PHOSPHATE ION, ... | | Authors: | Xin, B.G, Chen, W.F, Rety, S, Dai, Y.X, Xi, X.G. | | Deposit date: | 2018-08-20 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.019 Å) | | Cite: | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6ACO
 
 | |
6ACE
 
 | |
3R1K
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis H37Rv in complex with CoA and an acetamide moiety | | Descriptor: | ACETAMIDE, COENZYME A, Enhanced intracellular survival protein | | Authors: | Biswas, T, Chen, W, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7CPR
 
 | | glutamine synthetase from Drosophila | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase 2 cytoplasmic | | Authors: | Yin, H.S, Chen, W.T. | | Deposit date: | 2020-08-07 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Insight into the Contributions of the N-Terminus and Key Active-Site Residues to the Catalytic Efficiency of Glutamine Synthetase 2.
Biomolecules, 10, 2020
|
|
7EY7
 
 | | bacteriophage T7 tail complex | | Descriptor: | Internal virion protein gp14, Tail fiber protein, Tail tubular protein gp11, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY9
 
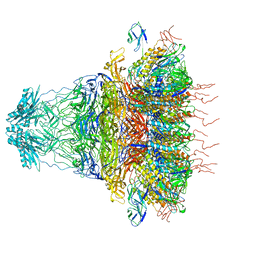 | | tail proteins | | Descriptor: | Tail fiber protein, Tail tubular protein gp11, Tail tubular protein gp12 | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2AFX
 
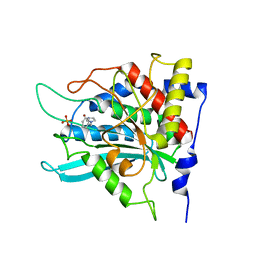 | | Crystal structure of human glutaminyl cyclase in complex with 1-benzylimidazole | | Descriptor: | 1-BENZYL-1H-IMIDAZOLE, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFO
 
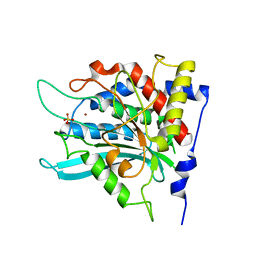 | | Crystal structure of human glutaminyl cyclase at pH 8.0 | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFW
 
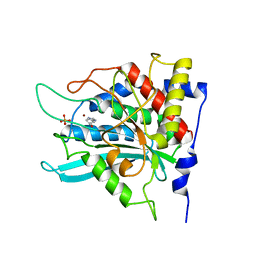 | | Crystal structure of human glutaminyl cyclase in complex with N-acetylhistamine | | Descriptor: | Glutaminyl-peptide cyclotransferase, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFZ
 
 | | Crystal structure of human glutaminyl cyclase in complex with 1-vinylimidazole | | Descriptor: | 1-VINYLIMIDAZOLE, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFS
 
 | | Crystal structure of the genetic mutant R54W of human glutaminyl cyclase | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFU
 
 | | Crystal structure of human glutaminyl cyclase in complex with glutamine t-butyl ester | | Descriptor: | Glutaminyl-peptide cyclotransferase, TERT-BUTYL D-ALPHA-GLUTAMINATE, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7EYB
 
 | | core proteins | | Descriptor: | Internal virion protein gp14, Internal virion protein gp15, Peptidoglycan transglycosylase gp16 | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY8
 
 | | portal | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY6
 
 | | The portal protein (GP8) of bacteriophage T7 | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FH5
 
 | | Structure of AdaV | | Descriptor: | AdaV, CHLORIDE ION, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-07-29 | | Release date: | 2022-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7EN6
 
 | | The crystal structure of Escherichia coli MurR in apo form | | Descriptor: | HTH-type transcriptional regulator MurR, PHOSPHATE ION | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7EN5
 
 | | The crystal structure of Escherichia coli MurR in complex with N-acetylglucosamine-6-phosphate | | Descriptor: | 2-METHOXYETHANOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7EN7
 
 | | The crystal structure of Escherichia coli MurR in complex with N-acetylmuramic-acid-6-phosphate | | Descriptor: | (2R)-2-[(2R,3R,4R,5S,6R)-3-acetamido-2,5-bis(oxidanyl)-6-(phosphonooxymethyl)oxan-4-yl]oxypropanoic acid, HTH-type transcriptional regulator MurR | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
6M31
 
 | |
8YVV
 
 | | The Crystal Structure of BTK from Biortus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of BTK from Biortus.
To Be Published
|
|
