1XK8
 
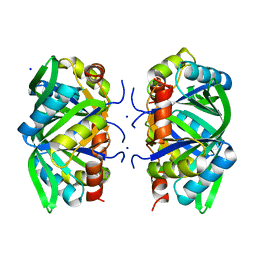 | | Divalent cation tolerant protein CUTA from Homo sapiens O60888 | | Descriptor: | Divalent cation tolerant protein CUTA, SODIUM ION | | Authors: | Tempel, W, Chen, L, Liu, Z.-J, Lee, D, Shah, A, Dailey, T.A, Mayer, M.R, Arendall III, W.B, Rose, J.P, Dailey, H.A, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-27 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Divalent cation tolerant protein CUTA from Homo sapiens O60888
To be published
|
|
1X6N
 
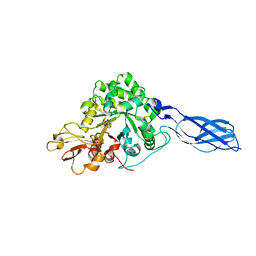 | | Crystal structure of S. marcescens chitinase A mutant W167A in complex with allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Chitinase A | | Authors: | Aronson Jr, N.N, Halloran, B.A, Alexyev, M.F, Zhou, X.E, Wang, Y, Meehan, E.J, Chen, L. | | Deposit date: | 2004-08-11 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Muation of Trp167 at the -3 subsite of the chitin-binding cleft of S. marcescens chitinase A causes enhanced transglycosylation
To be Published
|
|
1XE1
 
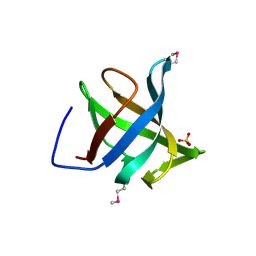 | | Hypothetical Protein From Pyrococcus Furiosus Pfu-880080-001 | | Descriptor: | SULFATE ION, hypothetical protein PF0907 | | Authors: | Chang, J.C, Zhao, M, Zhou, W, Liu, Z.J, Tempel, W, Arendall III, W.B, Chen, L, Lee, D, Habel, J.E, Rose, J.P, Richardson, J.S, Richardson, D.C, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-08 | | Release date: | 2004-09-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein From Pyrococcus Furiosus Pfu-880080-001
To be published
|
|
1XI6
 
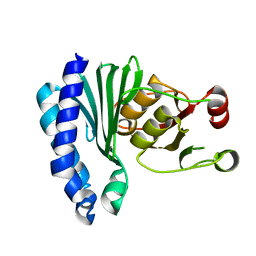 | | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001 | | Descriptor: | extragenic suppressor | | Authors: | Zhao, M, Chang, J.C, Zhou, W, Chen, L, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001
To be published
|
|
5XP5
 
 | | C-Src in complex with ATP-Chf | | Descriptor: | MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[(S)-fluoranyl-[oxidanyl(phosphonooxy)phosphoryl]methyl]phosphinic acid | | Authors: | Duan, Y, Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
1S9K
 
 | |
1RYQ
 
 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1SEN
 
 | | Endoplasmic reticulum protein Rp19 O95881 | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, Thioredoxin-like protein p19 | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Lee, D, Dailey, T.A, Mayer, M.R, Rose, J.P, Richardson, D.C, Richardson, J.S, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-07-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Endoplasmic reticulum protein Rp19
To be Published
|
|
2O94
 
 | |
1RD6
 
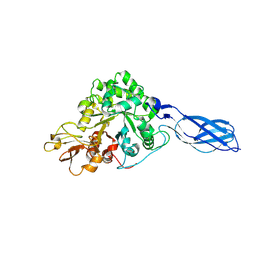 | | Crystal Structure of S. Marcescens Chitinase A Mutant W167A | | Descriptor: | Chitinase A | | Authors: | Aronson, N.N, Halloran, B.A, Alexyev, M.F, Zhou, X.E, Wang, Y, Meehan, E.J, Chen, L. | | Deposit date: | 2003-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutation of a conserved tryptophan in the chitin-binding cleft of Serratia marcescens chitinase A enhances transglycosylation.
Biosci.Biotechnol.Biochem., 70, 2006
|
|
1DK4
 
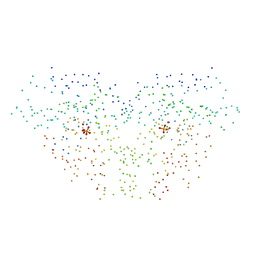 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | Descriptor: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | Deposit date: | 1999-12-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
1NLM
 
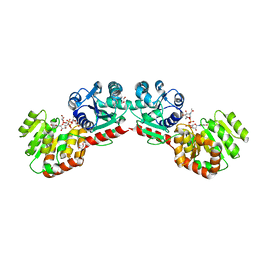 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | Descriptor: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8B9P
 
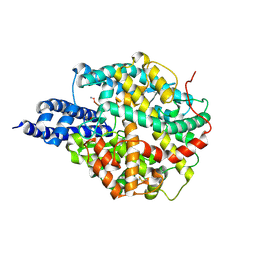 | | ACE2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-GLY-ARG-GLN-PHE-CYS-HIS-THR-LEU-MET-PRO-ARG-HIS-LEU-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-06 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
8BFW
 
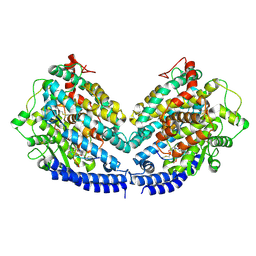 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-27 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
2O93
 
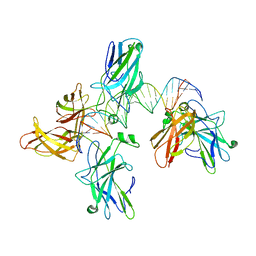 | |
8SV7
 
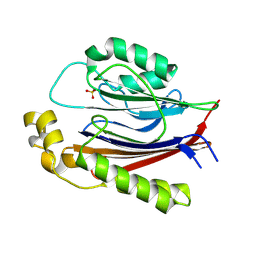 | |
4BM5
 
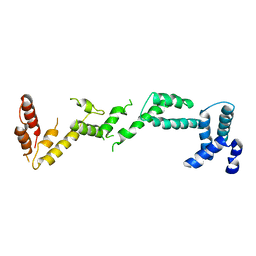 | | Chloroplast inner membrane protein TIC110 | | Descriptor: | SIMILAR TO CHLOROPLAST INNER MEMBRANE PROTEIN TIC110 | | Authors: | Tsai, J.-Y, Chu, C.-C, Yeh, Y.-H, Chen, L.-J, Li, H.-m, Hsiao, C.-D. | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural Characterizations of Chloroplast Translocon Protein Tic110.
Plant J., 75, 2013
|
|
2YX5
 
 | | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway | | Descriptor: | UPF0062 protein MJ1593 | | Authors: | Kanagawa, M, Baba, S, Agari, Y, Chen, L.Q, Fu, Z.-Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
2YXM
 
 | | Crystal structure of I-set domain of human Myosin Binding ProteinC | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Kishishita, S, Ohsawa, N, Murayama, K, Chen, L, Liu, Z, Terada, T, Shirouzu, M, Wang, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-26 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of I-set domain of human Myosin Binding ProteinC
To be Published
|
|
2YZ8
 
 | | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556) | | Descriptor: | Obscurin | | Authors: | Saijo, S, Ohsawa, N, Nishino, A, Kishishita, S, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556)
To be Published
|
|
1NA6
 
 | | Crystal structure of restriction endonuclease EcoRII mutant R88A | | Descriptor: | Restriction endonuclease EcoRII | | Authors: | Zhou, X.E, Wang, Y, Reuter, M, Mucke, M, Kruger, D.H, Meehan, E.J, Chen, L. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold.
J.Mol.Biol., 335, 2004
|
|
2YYO
 
 | | Crystal structure of human SPRY domain | | Descriptor: | SPRY domain-containing protein 3 | | Authors: | Kishishita, S, Uchikubo-Kamo, T, Murayama, K, Terada, T, Chen, L, Fu, Z.Q, Chrzas, J, Shirouzu, M, Wang, B.C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-30 | | Release date: | 2008-05-06 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human SPRY domain
To be Published
|
|
2Z0M
 
 | | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii | | Descriptor: | 337aa long hypothetical ATP-dependent RNA helicase deaD | | Authors: | Nakagawa, N, Kusano, S, Shirouzu, M, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii
To be Published
|
|
2YVI
 
 | | Crystal structure of a death domain of human ankryn protein | | Descriptor: | Ankyrin-1, GLYCEROL | | Authors: | Ihsanawati, Bessho, Y, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-12 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of a death domain of human ankryn protein
To be Published
|
|
2ZBV
 
 | | Crystal structure of uncharacterized conserved protein from Thermotoga maritima | | Descriptor: | ADENOSINE, Uncharacterized conserved protein | | Authors: | Ebihara, A, Fujimoto, Y, Kagawa, W, Fujikawa, N, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Thermotoga maritima
To be Published
|
|
