4M6B
 
 | | Crystal structure of yeast Swr1-Z domain in complex with H2A.Z-H2B dimer | | Descriptor: | Chimera protein of Histone H2B.1 and Histone H2A.Z, Helicase SWR1 | | Authors: | Hong, J.J, Feng, H.Q, Wang, F, Ranjan, A, Chen, J.H, Jiang, J.S, Girlando, R, Xiao, T.S, Wu, C, Bai, Y.W. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer.
Mol.Cell, 53, 2014
|
|
3OOR
 
 | | I-SceI mutant (K86R/G100T)complexed with C/G+4 DNA substrate | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | Authors: | Joshi, R, Chen, J.-H, Golden, B.L, Gimble, F.S. | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of I-SceI Homing Endonucleases with Increased DNA Recognition Site Specificity.
J.Mol.Biol., 405, 2011
|
|
3OOL
 
 | | I-SceI complexed with C/G+4 DNA substrate | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | Authors: | Joshi, R, Chen, J.-H, Golden, B.L, Gimble, F.S. | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of I-SceI Homing Endonucleases with Increased DNA Recognition Site Specificity.
J.Mol.Biol., 405, 2011
|
|
3P30
 
 | | crystal structure of the cluster II Fab 1281 in complex with HIV-1 gp41 ectodomain | | Descriptor: | 1281 Fab heavy chain, 1281 Fab light chain, HIV-1 gp41 | | Authors: | Frey, G, Chen, J, Rits-Volloch, S, Freeman, M.M, Zolla-Pazner, S, Chen, B. | | Deposit date: | 2010-10-04 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5YY6
 
 | | Crystal structure of Arabidopsis thaliana HPPD truncated mutant complexed with Benquitrione | | Descriptor: | 3-(2,6-dimethylphenyl)-1-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, W.C, Chen, J.N, Yang, G.F. | | Deposit date: | 2017-12-08 | | Release date: | 2019-01-16 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal Structure of 4-Hydroxyphenylpyruvate Dioxygenase in Complex with Substrate Reveals a New Starting Point for Herbicide Discovery.
Research (Wash D C), 2019, 2019
|
|
3WL5
 
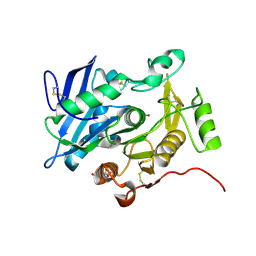 | | Crystal structure of pOPH S172C | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-07 | | Release date: | 2014-09-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
3WL8
 
 | | Crystal Structure of pOPH S172A with octanoic acid | | Descriptor: | CITRIC ACID, OCTANOIC ACID (CAPRYLIC ACID), Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
3WL7
 
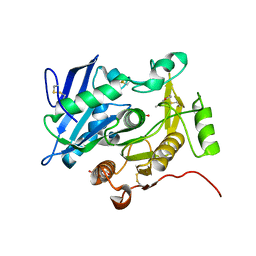 | | The complex structure of pOPH S172C with ligand, ACA | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase, pentane-2,4-dione | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
3WL6
 
 | | Crystal Structure of pOPH Native | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
3R5N
 
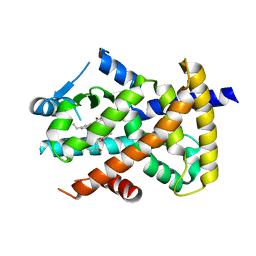 | | Crystal structure of PPARgammaLBD complexed with the agonist magnolol | | Descriptor: | 5,5'-di(prop-2-en-1-yl)biphenyl-2,2'-diol, Peroxisome proliferator-activated receptor gamma | | Authors: | Zhang, H, Chen, L, Chen, J, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular determinants of magnolol targeting both RXR(alpha) and PPAR(gamma).
Plos One, 6, 2011
|
|
3BYC
 
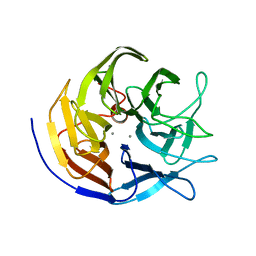 | | Joint neutron and X-ray structure of diisopropyl fluorophosphatase. Deuterium occupancies are 1-Q, where Q is occupancy of H | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Blum, M.-M, Mustyakimov, M, Ruterjans, H, Schoenborn, B.P, Langan, P, Chen, J.C.-H. | | Deposit date: | 2008-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Rapid determination of hydrogen positions and protonation states of diisopropyl fluorophosphatase by joint neutron and X-ray diffraction refinement.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3WWE
 
 | | The complex of pOPH with PEG | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chen, J, Guo, R.T, Du, G.C. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Roles of tryptophan residue and disulfide bond in the variable lid region of oxidized polyvinyl alcohol hydrolase
Biochem.Biophys.Res.Commun., 452, 2014
|
|
3WP0
 
 | | Crystal structure of Dlg GK in complex with a phosphor-Lgl2 peptide | | Descriptor: | Disks large homolog 4, GLYCEROL, Lethal(2) giant larvae protein homolog 2 | | Authors: | Zhu, J, Shang, Y, Wan, Q, Xia, Y, Chen, J, Du, Q, Zhang, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl
Cell Res., 24, 2014
|
|
1M4M
 
 | | Mouse Survivin | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, ZINC ION | | Authors: | Muchmore, S.W, Chen, J, Jakob, C, Zakula, D, Matayoshi, E.D, Wu, W, Zhang, H, Li, F, Ng, S.C, Altieri, D.C. | | Deposit date: | 2002-07-03 | | Release date: | 2002-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE AND MUTAGENIC ANALYSIS OF THE INHIBITOR-OF-APOPTOSIS PROTEIN SURVIVIN
MOL.CELL, 6, 2000
|
|
3WWD
 
 | | The complex of pOPH_S172C with DMSO | | Descriptor: | CITRIC ACID, DIMETHYL SULFOXIDE, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chen, J, Guo, R.T, Du, G.C. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Roles of tryptophan residue and disulfide bond in the variable lid region of oxidized polyvinyl alcohol hydrolase
Biochem.Biophys.Res.Commun., 452, 2014
|
|
3WWC
 
 | | The complex of pOPH_S172A of pNPB | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase, butanoic acid | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chen, J, Guo, R.T, Du, G.C. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Roles of tryptophan residue and disulfide bond in the variable lid region of oxidized polyvinyl alcohol hydrolase
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4FC1
 
 | |
3NAW
 
 | | Crystal structure of E. coli O157:H7 effector protein NleL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Lin, D.Y, Chen, J. | | Deposit date: | 2010-06-02 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and Structural Studies of a HECT-like Ubiquitin Ligase from Escherichia coli O157:H7.
J.Biol.Chem., 286, 2011
|
|
4AU6
 
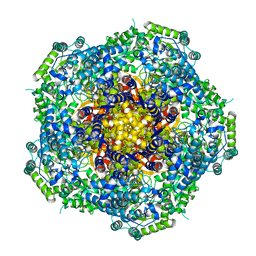 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Location of the Dsrna-Dependent Polymerase, Vp1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
4YL9
 
 | | Crystal Structure of wild-type of hsp14.1 from Sulfolobus solfatataricus P2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Heat shock protein Hsp20 | | Authors: | Liu, L, Chen, J.Y, Yun, C.H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Active-State Structures of a Small Heat-Shock Protein Revealed a Molecular Switch for Chaperone Function
Structure, 23, 2015
|
|
4YLC
 
 | |
4YLB
 
 | |
3C6R
 
 | | Low pH Immature Dengue Virus | | Descriptor: | Envelope protein, Peptide pr | | Authors: | Yu, I, Zhang, W, Holdway, H.A, Li, L, Kostyuchenko, V.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G, Chen, J. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Structure of the immature dengue virus at low pH primes proteolytic maturation
Science, 319, 2008
|
|
6C06
 
 | | Mycobacterium tuberculosis RNAP Holo/RbpA/Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
6C05
 
 | | Mycobacterium tuberculosis RNAP Holo/RbpA in relaxed state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
