2Q0O
 
 | | Crystal structure of an anti-activation complex in bacterial quorum sensing | | 分子名称: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, Probable transcriptional activator protein traR, Probable transcriptional repressor traM | | 著者 | Chen, G, Jeffrey, P.D, Fuqua, C, Shi, Y, Chen, L. | | 登録日 | 2007-05-22 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for antiactivation in bacterial quorum sensing.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5KYJ
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | 分子名称: | (6~{R})-5-(5-fluoranyl-2-methoxy-pyrimidin-4-yl)-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazole, Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta | | 著者 | Chen, G, McKeever, B.M. | | 登録日 | 2016-07-21 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5KYA
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | 分子名称: | Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta, [2-[(6~{R})-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazol-5-yl]-4-(trifluoromethyl)pyrimidin-5-yl]methanol | | 著者 | Chen, G, McKeever, B.M. | | 登録日 | 2016-07-21 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1RFY
 
 | | Crystal Structure of Quorum-Sensing Antiactivator TraM | | 分子名称: | Transcriptional repressor traM | | 著者 | Chen, G, Malenkos, J.W, Cha, M.R, Fuqua, C, Chen, L. | | 登録日 | 2003-11-10 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR
Mol.Microbiol., 52, 2004
|
|
7BW0
 
 | | Active human TGR5 complex with a synthetic agonist 23H | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Chen, G, Wang, X.K, Chen, Q, Hu, H.L, Ren, R.B. | | 登録日 | 2020-04-12 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of activated bile acids receptor TGR5 in complex with stimulatory G protein.
Signal Transduct Target Ther, 5, 2020
|
|
3BJC
 
 | | Crystal structure of the PDE5A catalytic domain in complex with a novel inhibitor | | 分子名称: | 5-ethoxy-4-(1-methyl-7-oxo-3-propyl-6,7-dihydro-1H-pyrazolo[4,3-d]pyrimidin-5-yl)thiophene-2-sulfonamide, MAGNESIUM ION, ZINC ION, ... | | 著者 | Chen, G, Wang, H, Howard, R, Cai, J, Wan, Y, Ke, H. | | 登録日 | 2007-12-03 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An insight into the pharmacophores of phosphodiesterase-5 inhibitors from synthetic and crystal structural studies
BIOCHEM.PHARM., 75, 2008
|
|
2DD1
 
 | | Three consecutive sheared GA pairs in 5'GGUGGAGGCU/3'PCCAAAGCCG | | 分子名称: | 5'-R(*GP*CP*CP*GP*AP*AP*AP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*GP*AP*GP*GP*CP*U)-3' | | 著者 | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | 登録日 | 2006-01-19 | | 公開日 | 2006-06-13 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
2DD3
 
 | | An alternating sheared AA pair in 5'GGUGAAGGCU/3'PCCGAAGCCG: II. The minor conformation with A6/A5/A16 stack | | 分子名称: | 5'-R(*GP*CP*CP*GP*AP*AP*GP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*AP*AP*GP*GP*CP*U)-3' | | 著者 | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | 登録日 | 2006-01-19 | | 公開日 | 2006-06-13 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
2DD2
 
 | | An alternating sheared AA pair in 5'GGUGAAGGCU/3'PCCGAAGCCG: I. The major conformation with A6/A15/A16 stack | | 分子名称: | 5'-R(*GP*CP*CP*GP*AP*AP*GP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*AP*AP*GP*GP*CP*U)-3' | | 著者 | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | 登録日 | 2006-01-19 | | 公開日 | 2006-06-13 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
1XV0
 
 | | Solution NMR structure of RNA internal loop with three consecutive sheared GA pairs in 5'GGUGGAGGCU/3'PCCGAAGCCG | | 分子名称: | 5'-R(*GP*CP*CP*GP*AP*AP*GP*CP*CP*(P5P)-3', 5'-R(*GP*GP*UP*GP*GP*AP*GP*GP*CP*U)-3' | | 著者 | Chen, G, Znosko, B.M, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | 登録日 | 2004-10-26 | | 公開日 | 2004-11-09 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of an RNA Internal Loop with Three Consecutive Sheared GA Pairs
Biochemistry, 44, 2005
|
|
5HCV
 
 | |
5I4V
 
 | | Discovery of novel, orally efficacious Liver X Receptor (LXR) beta agonists | | 分子名称: | Oxysterols receptor LXR-beta,Nuclear receptor coactivator 2, Retinoic acid receptor RXR-beta,Nuclear receptor coactivator 2, {2-[(2R)-4-[4-(hydroxymethyl)-3-(methylsulfonyl)phenyl]-2-(propan-2-yl)piperazin-1-yl]-4-(trifluoromethyl)pyrimidin-5-yl}methanol | | 著者 | Chen, G, McKeever, B.M. | | 登録日 | 2016-02-12 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Discovery of a Novel, Orally Efficacious Liver X Receptor (LXR) beta Agonist.
J.Med.Chem., 59, 2016
|
|
6KMJ
 
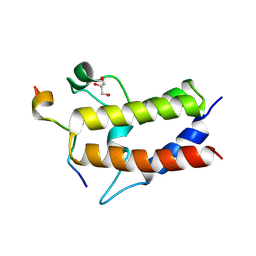 | | Crystal structure of Sth1 bromodomain in complex with H3K14Ac | | 分子名称: | GLYCEROL, Histone H3, Nuclear protein STH1/NPS1 | | 著者 | Chen, G, Li, W, Yan, F, Wang, D, Chen, Y. | | 登録日 | 2019-07-31 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex.
Structure, 28, 2020
|
|
6KMB
 
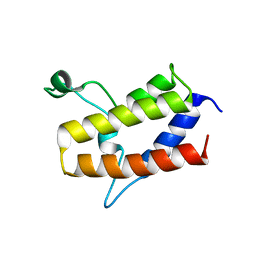 | | Crystal structure of Sth1 bromodomain | | 分子名称: | GLYCEROL, Nuclear protein STH1/NPS1 | | 著者 | Chen, G, Li, W, Yan, F, Wang, D, Chen, Y. | | 登録日 | 2019-07-31 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex.
Structure, 28, 2020
|
|
3QP4
 
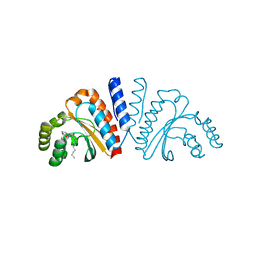 | | Crystal structure of CviR ligand-binding domain bound to C10-HSL | | 分子名称: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP2
 
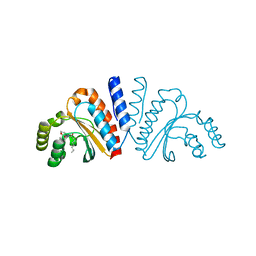 | | Crystal structure of CviR ligand-binding domain bound to C8-HSL | | 分子名称: | CviR transcriptional regulator, N-(2-OXOTETRAHYDROFURAN-3-YL)OCTANAMIDE | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.638 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP8
 
 | | Crystal structure of CviR (Chromobacterium violaceum 12472) ligand-binding domain bound to C10-HSL | | 分子名称: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP5
 
 | | Crystal structure of CviR bound to antagonist chlorolactone (CL) | | 分子名称: | 4-(4-chlorophenoxy)-N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, CviR transcriptional regulator | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.249 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP6
 
 | | Crystal structure of CviR (Chromobacterium violaceum 12472) bound to C6-HSL | | 分子名称: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP1
 
 | | Crystal structure of CviR ligand-binding domain bound to the native ligand C6-HSL | | 分子名称: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
2O81
 
 | |
6KAG
 
 | | Crystal structure of the SMARCB1/SMARCC2 subcomplex | | 分子名称: | SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | 著者 | Chen, G, Zhou, H, Giancotti, F.G, Long, J. | | 登録日 | 2019-06-22 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | A heterotrimeric SMARCB1-SMARCC2 subcomplex is required for the assembly and tumor suppression function of the BAF chromatin-remodeling complex.
Cell Discov, 6, 2020
|
|
7YKD
 
 | | Cryo-EM structure of the human chemerin receptor 1 complex with the C-terminal nonapeptide of chemerin | | 分子名称: | CHOLESTEROL, Chemerin-like receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Chen, G, Liao, Q, Ye, R.D, Wang, J. | | 登録日 | 2022-07-22 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Cryo-EM structure of the human chemerin receptor 1-Gi protein complex bound to the C-terminal nonapeptide of chemerin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1MK3
 
 | | SOLUTION STRUCTURE OF HUMAN BCL-W PROTEIN | | 分子名称: | Apoptosis regulator Bcl-W | | 著者 | Denisov, A.Y, Madiraju, M.S, Chen, G, Khadir, A, Beauparlant, P, Attardo, G, Shore, G.C, Gehring, K. | | 登録日 | 2002-08-28 | | 公開日 | 2003-06-03 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of human BCL-w: modulation of ligand binding by the C-terminal helix
J.BIOL.CHEM., 278, 2003
|
|
1F08
 
 | | CRYSTAL STRUCTURE OF THE DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN E1 FROM PAPILLOMAVIRUS | | 分子名称: | BROMIDE ION, REPLICATION PROTEIN E1 | | 著者 | Enemark, E.J, Chen, G, Vaughn, D.E, Stenlund, A, Joshua-Tor, L. | | 登録日 | 2000-05-15 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus.
Mol.Cell, 6, 2000
|
|
