8D56
 
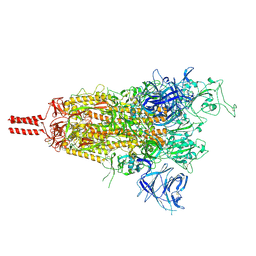 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7SBQ
 
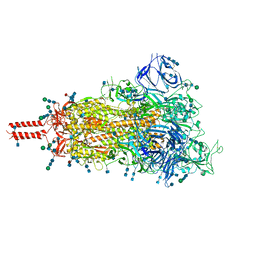 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBR
 
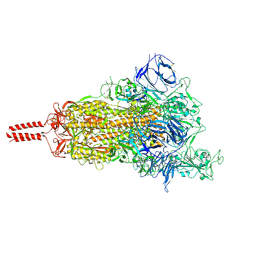 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBO
 
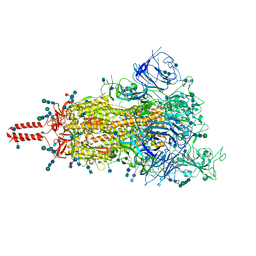 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBP
 
 | | Closed state of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBT
 
 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBK
 
 | | Closed state of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBS
 
 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBL
 
 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
6PWU
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Chen, B, Harrison, S.C. | | Deposit date: | 2019-07-23 | | Release date: | 2020-02-26 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
6XR8
 
 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
7TNW
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2022-01-21 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
7TO4
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2022-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
5C5B
 
 | | Crystal Structure of Human APPL BAR-PH Heterodimer | | Descriptor: | DCC-interacting protein 13-alpha, DCC-interacting protein 13-beta, GLYCEROL | | Authors: | Chen, Y.J, Chen, B. | | Deposit date: | 2015-06-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human APPL BAR-PH Heterodimer
To Be Published
|
|
6XRA
 
 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Rits-Volloch, S, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
6V4T
 
 | | MPER-TMD of HIV-1 Env bound with the entry inhibitor S2C3 | | Descriptor: | 4,4'-(decane-1,10-diyl)bis(9-amino-2,3-dihydro-1H-cyclopenta[b]quinolin-4-ium), Envelope glycoprotein gp160 | | Authors: | Xiao, T, Frey, G, Fu, Q, Lavine, C.L, Scott, D.A, Seaman, M.S, Chou, J.J, Chen, B. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes.
Nat.Chem.Biol., 16, 2020
|
|
6OSK
 
 | | RF1 accommodated 70S complex at 60 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSQ
 
 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-26 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OUO
 
 | | RF2 accommodated state bound 70S complex at long incubation time | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
7CBI
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)pentyl (2~{S},3~{R})-2-azanyl-3-oxidanyl-butanoate, GLYCEROL, ... | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
3UA0
 
 | | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease | | Descriptor: | Fibroin heavy chain | | Authors: | He, Y.-X, Zhang, N.-N, Chen, B.-Y, Li, W.-F, Chen, Y.-X, Zhou, C.-Z. | | Deposit date: | 2011-10-20 | | Release date: | 2012-03-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease.
J.Mol.Biol., 418, 2012
|
|
1ZO4
 
 | | Crystal Structure Of A328S Mutant Of The Heme Domain Of P450BM-3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450:NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Hegda, A, Chen, B, Haines, D.C, Bondlela, M, Mullin, D, Graham, S.E, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2005-05-12 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A single active-site mutation of P450BM-3 dramatically enhances substrate binding and rate of product formation.
Biochemistry, 50, 2011
|
|
3VPM
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
3VPO
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
7CBG
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-N-[(E)-4-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]but-2-enyl]-2-(methylamino)-3-oxidanyl-butanamide, Threonine--tRNA ligase, ZINC ION | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
