2NP5
 
 | | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1. | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Kagan, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1.
TO BE PUBLISHED
|
|
3F1B
 
 | | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, TetR-like transcriptional regulator | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3F6O
 
 | | Crystal structure of ArsR family transcriptional regulator, RHA00566 | | Descriptor: | BROMIDE ION, Probable transcriptional regulator, ArsR family protein | | Authors: | Dong, A, Xu, X, Zheng, H, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ArsR family transcriptional regulator, RHA00566
To be Published
|
|
4IC1
 
 | | Crystal structure of SSO0001 | | Descriptor: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-09 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
3F4A
 
 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
3P48
 
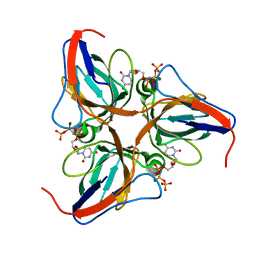 | | Structure of the yeast dUTPase DUT1 in complex with dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Petit, P, Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3OUT
 
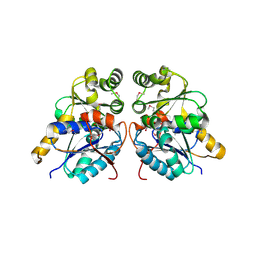 | | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate. | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Kudriska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-09-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
To be Published
|
|
3FBS
 
 | | The crystal structure of the oxidoreductase from Agrobacterium tumefaciens | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Oxidoreductase, ... | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the oxidoreductase from Agrobacterium tumefaciens
To be Published, 2008
|
|
3P52
 
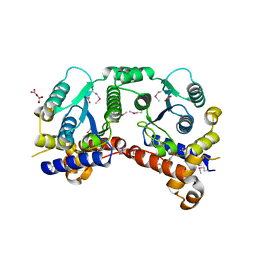 | | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NITRATE ION | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-07 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion
To be Published
|
|
6PNY
 
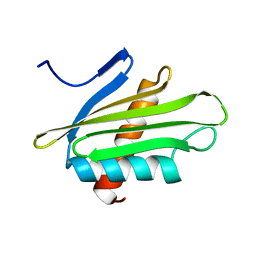 | | X-ray Structure of Flpp3 | | Descriptor: | Flpp3 | | Authors: | Zook, J.D, Shekhar, M, Hansen, D.T, Conrad, C, Grant, T.D, Gupta, C, White, T, Barty, A, Basu, S, Zhao, Y, Zatsepin, N.A, Ishchenko, A, Batyuk, A, Gati, C, Li, C, Galli, L, Coe, J, Hunter, M, Liang, M, Weierstall, U, Nelson, G, James, D, Stauch, B, Craciunescu, F, Thifault, D, Liu, W, Cherezov, V, Singharoy, A, Fromme, P. | | Deposit date: | 2019-07-03 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | XFEL and NMR Structures of Francisella Lipoprotein Reveal Conformational Space of Drug Target against Tularemia.
Structure, 28, 2020
|
|
3FOV
 
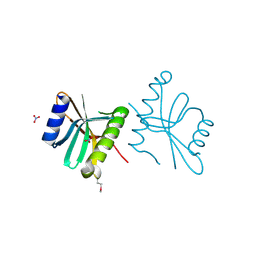 | | Crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris | | Descriptor: | NITRATE ION, UPF0102 protein RPA0323 | | Authors: | Osipiuk, J, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-02 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris.
To be Published
|
|
3F5R
 
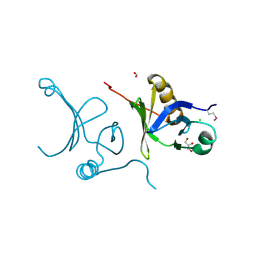 | | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p). | | Descriptor: | CHLORIDE ION, FACT complex subunit POB3, FORMIC ACID, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p).
To be Published
|
|
1P8C
 
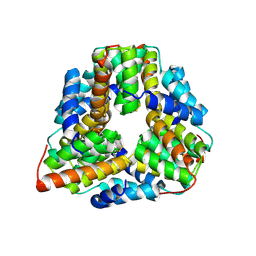 | | Crystal structure of TM1620 (APC4843) from Thermotoga maritima | | Descriptor: | conserved hypothetical protein | | Authors: | Kim, Y, Joachimiak, A, Brunzelle, J.S, Korolev, S.V, Edwards, A, Xu, X, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima protein TM1620 (APC4843)
To be Published
|
|
2OZV
 
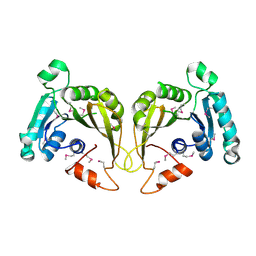 | | Crystal structure of a predicted O-methyltransferase, protein Atu636 from Agrobacterium tumefaciens. | | Descriptor: | Hypothetical protein Atu0636 | | Authors: | Cuff, M.E, Xu, X, Zheng, X, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a predicted O-methyltransferase, protein Atu636 from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
2O99
 
 | | The crystal structure of E.coli IclR C-terminal fragment in complex with glyoxylate | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, GLYCOLIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
2OFY
 
 | | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | Putative XRE-family transcriptional regulator | | Authors: | Shumilin, I.A, Skarina, T, Onopriyenko, O, Yim, V, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
2ODF
 
 | | The crystal structure of gene product Atu2144 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu2144, SULFATE ION | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of gene product Atu2144 from Agrobacterium tumefaciens
To be Published
|
|
3D3R
 
 | | Crystal structure of the hydrogenase assembly chaperone HypC/HupF family protein from Shewanella oneidensis MR-1 | | Descriptor: | Hydrogenase assembly chaperone hypC/hupF | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Hydrogenase Assembly Chaperone HypC/HupF Family Protein from Shewanella oneidensis MR-1.
To be Published
|
|
3MTJ
 
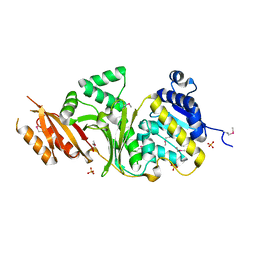 | | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A | | Descriptor: | Homoserine dehydrogenase, SULFATE ION | | Authors: | Stein, A.J, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A
To be Published
|
|
2O6B
 
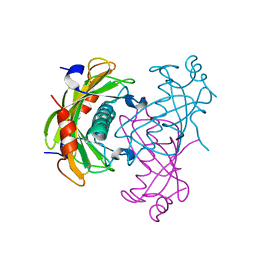 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-07 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2OV9
 
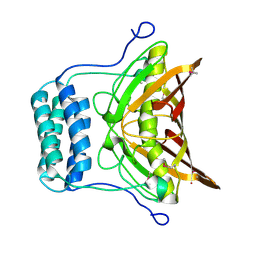 | | Crystal structure of protein RHA08564, thioesterase superfamily protein | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-13 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of protein RHA08564, thioesterase superfamily protein
To be Published
|
|
2DXQ
 
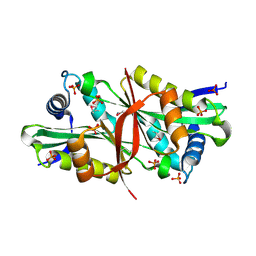 | | Putative acetyltransferase from Agrobacterium tumefaciens str. C58 | | Descriptor: | Acetyltransferase, PHOSPHATE ION | | Authors: | Binkowski, T.A, Xu, X, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Putative acetyltransferase from Agrobacterium tumefaciens str. C58
To be published
|
|
2PHN
 
 | | Crystal structure of an amide bond forming F420-gamma glutamyl ligase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, F420-0:gamma-glutamyl ligase, ... | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an Amide Bond Forming F(420):gammagamma-glutamyl Ligase from Archaeoglobus Fulgidus - A Member of a New Family of Non-ribosomal Peptide Synthases.
J.Mol.Biol., 372, 2007
|
|
1ORU
 
 | | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis | | Descriptor: | CHLORIDE ION, SULFATE ION, yuaD protein | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-15 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis
To be Published
|
|
1OTK
 
 | | Structural Genomics, Protein paaC | | Descriptor: | Phenylacetic acid degradation protein paaC | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-21 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2 A crystal structure of protein paaC from E. Coli
To be Published
|
|
