7MVX
 
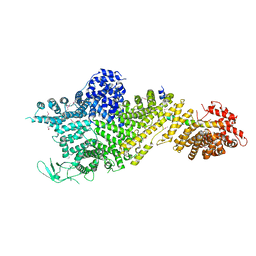 | | Crystal structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | 分子名称: | Nucleoporin NIC96, Nucleoporin NUP188 | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-05-15 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | X-RAY DIFFRACTION (4.35 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVY
 
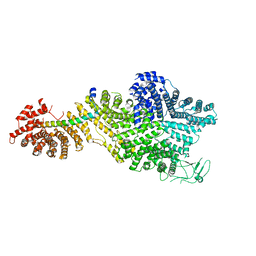 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | 分子名称: | Nucleoporin NIC96, Nucleoporin NUP188 | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-05-15 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.39 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MW1
 
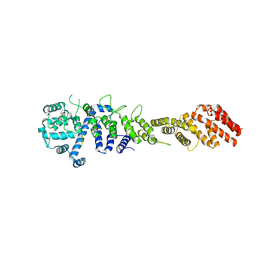 | | Crystal structure of the Homo sapiens NUP93-NUP53 complex (NUP93 residues 174-819; NUP53 residues 84-150) | | 分子名称: | Nuclear pore complex protein Nup93, Nucleoporin Nup35 | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-05-15 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MW0
 
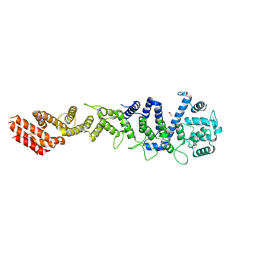 | | Crystal structure of Homo sapiens NUP93 solenoid (residues 174-819) | | 分子名称: | 1,2-ETHANEDIOL, Nuclear pore complex protein Nup93 | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-05-15 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
5EK9
 
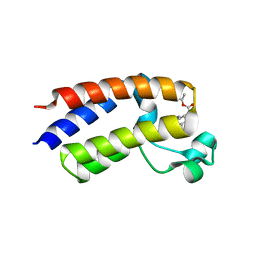 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline inhibitor | | 分子名称: | Bromodomain-containing protein 2, propan-2-yl ~{N}-[(2~{S},4~{R})-1-ethanoyl-6-(furan-2-yl)-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | 著者 | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Picaud, S, Fedorov, O, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-03 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Exploiting a water network to achieve enthalpy-driven, bromodomain-selective BET inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
2FM2
 
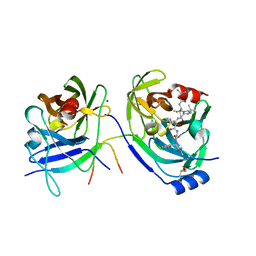 | | HCV NS3-4A protease domain complexed with a ketoamide inhibitor, SCH446211 | | 分子名称: | BETA-MERCAPTOETHANOL, NS3 protease/helicase, NS4a protein, ... | | 著者 | Yi, M, Tong, X, Skelton, A, Chase, R, Chen, T, Prongay, A, Bogen, S.L, Saksena, A.K, Njoroge, F.G, Veselenak, R.L, Pyles, R.B, Bourne, N, Malcolm, B.A, Lemon, S.M. | | 登録日 | 2006-01-06 | | 公開日 | 2006-02-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations
J.Biol.Chem., 281, 2006
|
|
6LHC
 
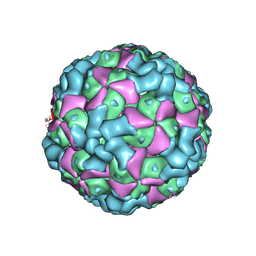 | | The cryo-EM structure of coxsackievirus A16 empty particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.43 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
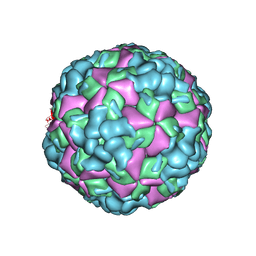 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | 分子名称: | VP1 protein, VP2 protein, VP3 protein | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHB
 
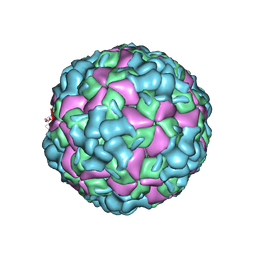 | | The cryo-EM structure of coxsackievirus A16 A-particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHP
 
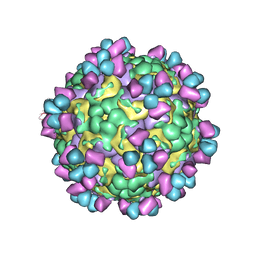 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 14B10 | | 分子名称: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
5JFG
 
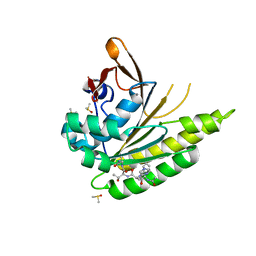 | | Structure of humanised RadA-mutant humRadA22F in complex with peptide FHTA | | 分子名称: | CALCIUM ION, DIMETHYL SULFOXIDE, DNA repair and recombination protein RadA, ... | | 著者 | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | 登録日 | 2016-04-19 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Humanisation of RadA from Pyrococcus furiosus
To Be Published
|
|
7BOB
 
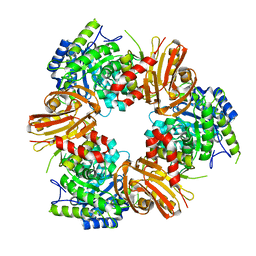 | | Exo-beta-1,4-mannosidase Op5Man5 from Opitutaceae bacterium strain TAV5 | | 分子名称: | Endo-beta-mannanase | | 著者 | Kalyani, D.C, Reichenbach, T, Keskitalo, M.M, Conrad, J, Aspeborg, H, Divne, C. | | 登録日 | 2021-01-24 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a homotrimeric verrucomicrobial exo - beta -1,4-mannosidase active in the hindgut of the wood-feeding termite Reticulitermes flavipes .
J Struct Biol X, 5, 2021
|
|
5X0R
 
 | | Crystal Structure of PXR LBD Complexed with SJB7 | | 分子名称: | 4-[(4-tert-butylphenyl)sulfonyl]-1-(2,4-dimethoxy-5-methylphenyl)-5-methyl-1H-1,2,3-triazole, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | 著者 | Lv, L, Lin, W, Chai, S.C, Zhang, Q, Chen, T. | | 登録日 | 2017-01-23 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.665 Å) | | 主引用文献 | SPA70 is a potent antagonist of human pregnane X receptor.
Nat Commun, 8, 2017
|
|
7BHS
 
 | | Crystal structure of MAT2a with quinazoline fragment 2 bound in the allosteric site | | 分子名称: | 6-chloranyl-2-methoxy-4-phenyl-quinazoline, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHU
 
 | | Crystal structure of MAT2a with elaborated fragment 26 bound in the allosteric site | | 分子名称: | 1,2-ETHANEDIOL, 7-chloranyl-4-(dimethylamino)-1-(2-hydroxyethyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, ... | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHW
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 29) | | 分子名称: | 7-chloranyl-4-(dimethylamino)-1-(3-methylphenyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHR
 
 | | Crystal structure of MAT2a with triazinone fragment 1 bound in the allosteric site | | 分子名称: | 4-(dimethylamino)-6-ethoxy-1~{H}-1,3,5-triazin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHV
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor and in vivo tool compound 28 | | 分子名称: | 7-chloranyl-4-(dimethylamino)-1-phenyl-quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHT
 
 | | Crystal structure of MAT2a with quinazolinone fragment 5 bound in the allosteric site | | 分子名称: | 7-chloranyl-4-(dimethylamino)-1~{H}-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.052 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHX
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 31) | | 分子名称: | 7-chloranyl-4-(dimethylamino)-1-pyridin-3-yl-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
6JXM
 
 | | Crystal Structure of phi29 pRNA domain II | | 分子名称: | BARIUM ION, MAGNESIUM ION, RNA (97-mer) | | 著者 | Cai, R, Price, I.R, Ding, F, Wu, F, Chen, T, Zhang, Y, Liu, G, Jardine, P.J, Lu, C, Ke, A. | | 登録日 | 2019-04-24 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | ATP/ADP modulates gp16-pRNA conformational change in the Phi29 DNA packaging motor.
Nucleic Acids Res., 47, 2019
|
|
1XR8
 
 | | Crystal Structures of HLA-B*1501 in Complex with Peptides from Human UbcH6 and Epstein-Barr Virus EBNA-3 | | 分子名称: | Beta-2-microglobulin, EBNA-3 nuclear protein, GLYCEROL, ... | | 著者 | Roder, G, Blicher, T, Johannessen, B.R, Kristensen, O, Buus, S, Gajhede, M. | | 登録日 | 2004-10-14 | | 公開日 | 2005-04-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of two peptide-HLA-B*1501 complexes; structural characterization of the HLA-B62 supertype
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1XR9
 
 | | Crystal Structures of HLA-B*1501 in Complex with Peptides from Human UbcH6 and Epstein-Barr Virus EBNA-3 | | 分子名称: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | 著者 | Roder, G, Blicher, T, Johannessen, B.R, Kristensen, O, Buus, S, Gajhede, M. | | 登録日 | 2004-10-14 | | 公開日 | 2005-04-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.788 Å) | | 主引用文献 | Crystal structures of two peptide-HLA-B*1501 complexes; structural characterization of the HLA-B62 supertype
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2L87
 
 | | The 27-residue N-terminus CCR5-peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide | | 分子名称: | C-C chemokine receptor type 5 | | 著者 | Schnur, E, Noah, E, Ayzenshtat, I, Sargsyan, H, Inui, T, Ding, F.X, Arshava, B, Sagi, Y, Kessler, N, Levy, R, Scherf, T, Naider, F, Anglister, J. | | 登録日 | 2011-01-06 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Conformation and Orientation of a 27-Residue CCR5 Peptide in a Ternary Complex with HIV-1 gp120 and a CD4-Mimic Peptide.
J.Mol.Biol., 410, 2011
|
|
1Y7W
 
 | | Crystal structure of a halotolerant carbonic anhydrase from Dunaliella salina | | 分子名称: | ACETIC ACID, Halotolerant alpha-type carbonic anhydrase (dCA II), SODIUM ION, ... | | 著者 | Premkumar, L, Greenblatt, H.M, Bageshwar, U.K, Savchenko, T, Gokhman, I, Sussman, J.L, Zamir, A, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2004-12-10 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Three-dimensional structure of a halotolerant algal carbonic anhydrase predicts halotolerance of a mammalian homolog.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
