1N87
 
 | |
8GEX
 
 | |
9PCY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED FRENCH BEAN PLASTOCYANIN AND COMPARISON WITH THE CRYSTAL STRUCTURE OF POPLAR PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Moore, J.M, Lepre, C.A, Gippert, G.P, Chazin, W.J, Case, D.A, Wright, P.E. | | Deposit date: | 1991-03-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced French bean plastocyanin and comparison with the crystal structure of poplar plastocyanin.
J.Mol.Biol., 221, 1991
|
|
8S9W
 
 | | Murine S100A7/S100A15 in presence of calcium | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Harrison, S.A, Naretto, A, Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-03-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Comparative analysis of the physical properties of murine and human S100A7: Insight into why zinc piracy is mediated by human but not murine S100A7.
J.Biol.Chem., 299, 2023
|
|
1HV2
 
 | | SOLUTION STRUCTURE OF YEAST ELONGIN C IN COMPLEX WITH A VON HIPPEL-LINDAU PEPTIDE | | Descriptor: | ELONGIN C, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR | | Authors: | Botuyan, M.V, Mer, G, Yi, G.-S, Koth, C.M, Case, D.A, Edwards, A.M, Chazin, W.J, Arrowsmith, C.H. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide.
J.Mol.Biol., 312, 2001
|
|
4O0A
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-{4-[({[4-(5-carboxyfuran-2-yl)-2-chlorophenyl]carbonothioyl}amino)methyl]phenyl}-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejniczak, E.T, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-12-13 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J. Med. Chem., 56, 2013
|
|
7LMW
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 7-methyl-3-(1~{H}-pyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
5I7M
 
 | |
7TL3
 
 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF431 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
7TL4
 
 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 6YF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
7TL2
 
 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF412 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, GLYCEROL, ... | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
2KWQ
 
 | | Mcm10 C-terminal DNA binding domain | | Descriptor: | Protein MCM10 homolog, ZINC ION | | Authors: | Robertson, P.D, Chagot, B, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif.
J.Biol.Chem., 285, 2010
|
|
7LML
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 6-iodanyl-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
2L53
 
 | |
3LRV
 
 | |
3L9Q
 
 | |
2CNP
 
 | |
3CJJ
 
 | | Crystal structure of human rage ligand-binding domain | | Descriptor: | ACETATE ION, Advanced glycosylation end product-specific receptor, ZINC ION | | Authors: | Koch, M, Dattilo, B.M, Schiefner, A, Diez, J, Chazin, W.J, Fritz, G. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand recognition and activation of RAGE.
Structure, 18, 2010
|
|
2BCB
 
 | |
2BCA
 
 | |
1JWD
 
 | |
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
1Z1D
 
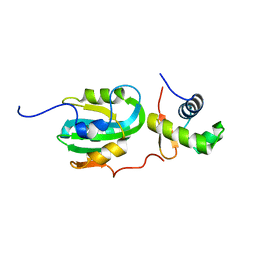 | | Structural Model for the interaction between RPA32 C-terminal domain and SV40 T antigen origin binding domain. | | Descriptor: | Large T antigen, Replication protein A 32 kDa subunit | | Authors: | Arunkumar, A.I, Klimovich, V, Jiang, X, Ott, R.D, Mizoue, L, Fanning, E, Chazin, W.J. | | Deposit date: | 2005-03-03 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome.
Nat.Struct.Mol.Biol., 12, 2005
|
|
8G9F
 
 | | Complete auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G9O
 
 | | Complete DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
