1LBU
 
 | | HYDROLASE METALLO (ZN) DD-PEPTIDASE | | Descriptor: | MURAMOYL-PENTAPEPTIDE CARBOXYPEPTIDASE, ZINC ION | | Authors: | Charlier, P, Wery, J.-P, Dideberg, O, Frere, J.-M. | | Deposit date: | 1996-03-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Streptomyces Albus G D-Ala-A-Ala Carboxypeptidase
Handbook of Metalloproteins, 3, 2004
|
|
4M3K
 
 | | Structure of a single domain camelid antibody fragment cAb-H7S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, CHLORIDE ION, Camelid heavy-chain antibody variable fragment cAb-H7S | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4D2O
 
 | | Crystal structure of the class A extended-spectrum beta-lactamase PER- 2 | | Descriptor: | PER-2 BETA-LACTAMASE | | Authors: | Power, P, Herman, R, Ruggiero, M, Kerff, F, Galleni, M, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2014-05-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Extended-Spectrum Beta-Lactamase Per- 2 and Insights Into the Role of Specific Residues in the Interaction with Beta-Lactams and Beta-Lactamase Inhibitors.
Antimicrob.Agents Chemother., 58, 2014
|
|
1K4F
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 AT 1.6 A RESOLUTION | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the class D beta-lactamase OXA-2
To be Published
|
|
1K6R
 
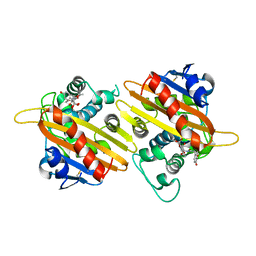 | | STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH MOXALACTAM | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, Beta-lactamase PSE-2 | | Authors: | Kerff, F, Fonze, E, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-17 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
To be Published
|
|
1K4E
 
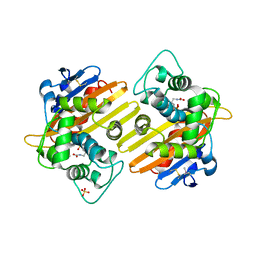 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASES OXA-10 DETERMINED BY MAD PHASING WITH SELENOMETHIONINE | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURE OF CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1K6S
 
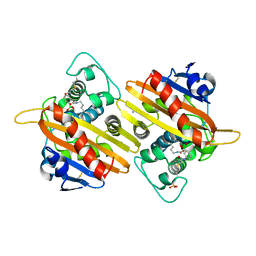 | | STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH A PHENYLBORONIC ACID | | Descriptor: | 4-IODO-ACETAMIDO PHENYLBORONIC ACID, Beta-lactamase PSE-2, CALCIUM ION, ... | | Authors: | Kerff, F, Fonze, E, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
To be Published
|
|
7AP7
 
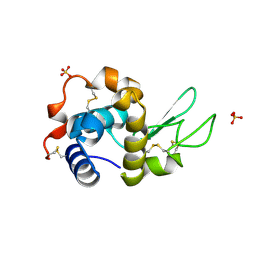 | | Structure of the W64R amyloidogenic variant of human lysozyme | | Descriptor: | Lysozyme C, SULFATE ION | | Authors: | Vettore, N, Herman, R, Kerff, F, Charlier, P, Sauvage, E, Brans, A, Morray, J, Dobson, C, Kumita, J, Dumoulin, M. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Characterisation of the structural, dynamic and aggregation properties of the W64R amyloidogenic variant of human lysozyme.
Biophys.Chem., 271, 2021
|
|
3TVL
 
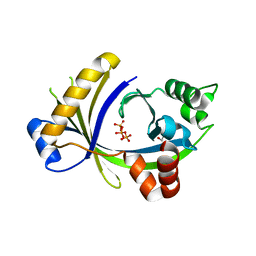 | | Complex between the human thiamine triphosphatase and triphosphate | | Descriptor: | 1,2-ETHANEDIOL, TRIPHOSPHATE, Thiamine-triphosphatase | | Authors: | Delvaux, D, Herman, R, Sauvage, E, Wins, P, Bettendorff, L, Charlier, P, Kerff, F. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of specificity and catalytic mechanism in mammalian 25-kDa thiamine triphosphatase.
Biochim.Biophys.Acta, 1830, 2013
|
|
4M3J
 
 | | Structure of a single-domain camelid antibody fragment cAb-H7S specific of the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Camelid heavy-chain antibody variable fragment cAb-H7S, SULFATE ION | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4N1H
 
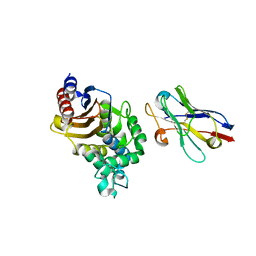 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
1SKF
 
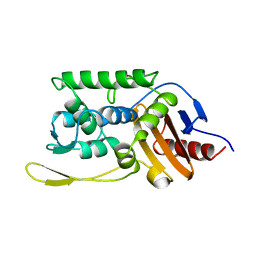 | | CRYSTAL STRUCTURE OF THE STREPTOMYCES K15 DD-TRANSPEPTIDASE | | Descriptor: | D-ALANYL-D-ALANINE TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 1998-08-20 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a penicilloyl-serine transferase of intermediate penicillin sensitivity. The DD-transpeptidase of streptomyces K15.
J.Biol.Chem., 274, 1999
|
|
5AEB
 
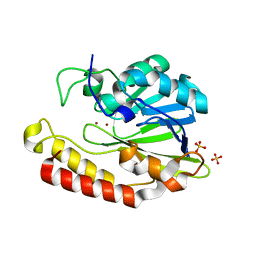 | | Crystal structure of the class B3 di-zinc metallo-beta-lactamase LRA- 12 from an Alaskan soil metagenome. | | Descriptor: | COBALT (II) ION, LRA-12, SULFATE ION, ... | | Authors: | Power, P, Herman, R, Kerff, F, Bouillenne, F, Rodriguez, M.M, Galleni, M, Handelsman, J, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2015-08-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and kinetic analysis of the class B3 di-zinc metallo-beta-lactamase LRA-12 from an Alaskan soil metagenome.
PLoS ONE, 12, 2017
|
|
1ES2
 
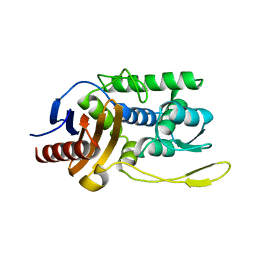 | | S96A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES3
 
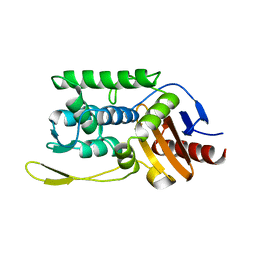 | | C98A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE, SODIUM ION | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ESI
 
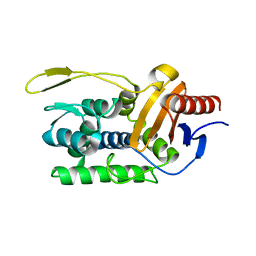 | |
1ES4
 
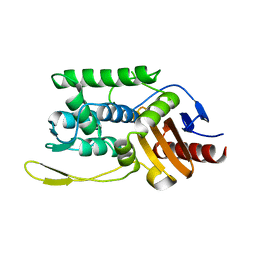 | | C98N mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES5
 
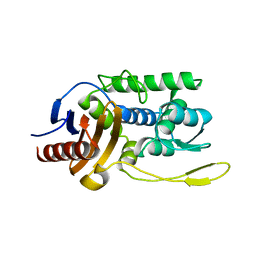 | |
2BH7
 
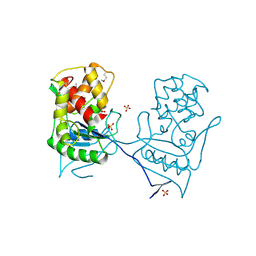 | | Crystal structure of a SeMet derivative of AmiD at 2.2 angstroms | | Descriptor: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE, SULFATE ION, ZINC ION | | Authors: | Petrella, S, Herman, R, Sauvage, E, Genereux, C, Pennartz, A, Joris, B, Charlier, P. | | Deposit date: | 2005-01-07 | | Release date: | 2006-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-L-Alanine Amidase Amid from Escherichia Coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
1J9M
 
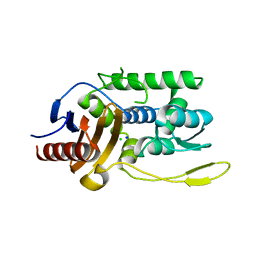 | | K38H mutant of Streptomyces K15 DD-transpeptidase | | Descriptor: | CHLORIDE ION, DD-transpeptidase, SODIUM ION | | Authors: | Fonze, E, Rhazi, N, Nguyen-Disteche, M, Charlier, P. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1NRF
 
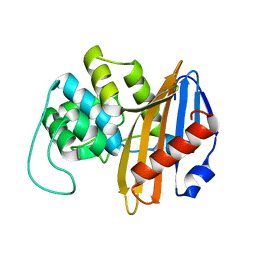 | | C-terminal domain of the Bacillus licheniformis BlaR penicillin-receptor | | Descriptor: | REGULATORY PROTEIN BLAR1 | | Authors: | Kerff, F, Charlier, P, Columbo, M.L, Sauvage, E, Brans, A, Frere, J.M, Joris, B, Fonze, E. | | Deposit date: | 2003-01-24 | | Release date: | 2004-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the sensor domain of the BlaR penicillin receptor from Bacillus licheniformis.
Biochemistry, 42, 2003
|
|
2CC1
 
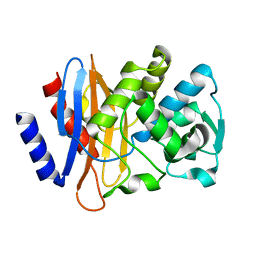 | |
6FLY
 
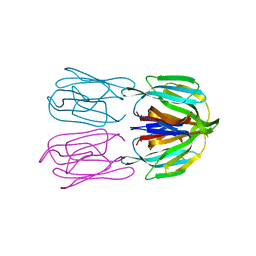 | | Structure of AcmJRL, a mannose binding jacalin related lectin from Ananas comosus, in complex with mannose. | | Descriptor: | Jacalin-like lectin, alpha-D-mannopyranose | | Authors: | Azarkan, M, Herman, R, El Mahyaoui, R, Sauvage, E, Vanden Broeck, A, Charlier, P. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem.
Sci Rep, 8, 2018
|
|
6FLZ
 
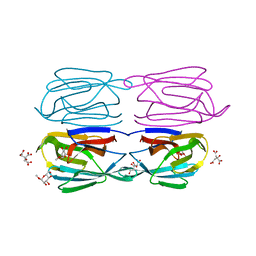 | | Structure of AcmJRL, a mannose binding jacalin related lectin from Ananas comosus, in complex with methyl-mannose. | | Descriptor: | CITRIC ACID, Jacalin-like lectin, methyl alpha-D-mannopyranoside | | Authors: | Azarkan, M, Herman, R, El Mahyaoui, R, Sauvage, E, Vanden Broeck, A, Charlier, P. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem.
Sci Rep, 8, 2018
|
|
6FLW
 
 | | Structure of AcmJRL, a mannose binding jacalin related lectin from Ananas comosus. | | Descriptor: | CITRIC ACID, Jacalin-like lectin | | Authors: | Azarkan, M, Herman, R, El Mahyaoui, R, Sauvage, E, Vanden Broeck, A, Charlier, P. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem.
Sci Rep, 8, 2018
|
|
