4M2C
 
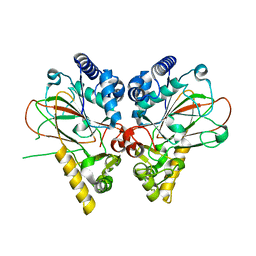 | | Crystal structure of non-heme iron oxygenase OrfP in complex with Fe and D-Arg | | Descriptor: | D-ARGININE, FE (III) ION, L-arginine beta-hydroxylase | | Authors: | Chang, C.Y, Liu, Y.C, Lyu, S.Y, Wu, C.C, Li, T.L. | | Deposit date: | 2013-08-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7X3N
 
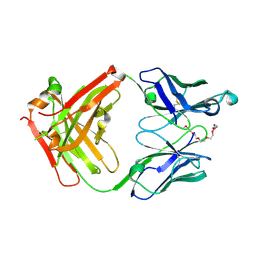 | | Crystal structure of anti-mPEG h15-2b Fab | | Descriptor: | 15-2b-H, 15-2b-L, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL | | Authors: | Chang, C.Y, Nguyen, T.M.T, Toh, S.I, Su, Y.C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural determination of an antibody that specifically recognizes polyethylene glycol with a terminal methoxy group
Commun Chem, 5, 2022
|
|
7Y0G
 
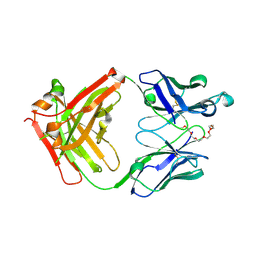 | | Crystal structure of anti-mPEG h15-2b Fab | | Descriptor: | 15-2b heavy chain, 15-2b light chain, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL | | Authors: | Chang, C.Y, Nguyen, T.M.T, Lin, E.C, Su, Y.C. | | Deposit date: | 2022-06-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural determination of an antibody that specifically recognizes polyethylene glycol with a terminal methoxy group.
Commun Chem, 5, 2022
|
|
7XKX
 
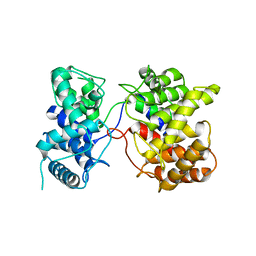 | | Crystal structure of Tpn2 | | Descriptor: | SQHop_cyclase_C domain-containing protein | | Authors: | Chang, C.Y, Stowell, E.A, Lin, Y.L, Ehrenberger, M.A, Rudolf, J.D. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure-guided product determination of the bacterial type II diterpene synthase Tpn2.
Commun Chem, 5, 2022
|
|
5UMY
 
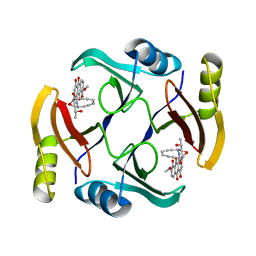 | | Crystal structure of TnmS3 in complex with tiancimycin | | Descriptor: | (1aS,11S,11aR,14Z,18R)-3,8,18-trihydroxy-11a-[(1R)-1-hydroxyethyl]-7-methoxy-11,11a-dihydro-4H-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinoline-4,9(10H)-dione, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMP
 
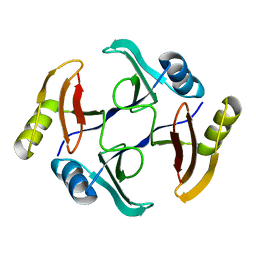 | | Crystal structure of TnmS3, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMW
 
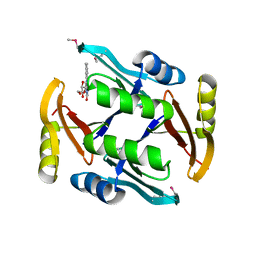 | | Crystal structure of TnmS2, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMQ
 
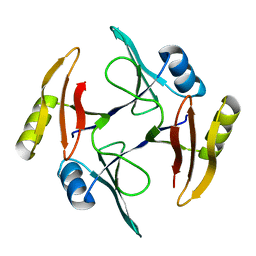 | | Crystal structure of TnmS1, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMX
 
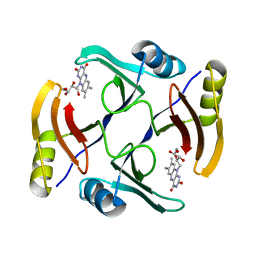 | | Crystal structure of TnmS3 in complex with riboflavin | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5W27
 
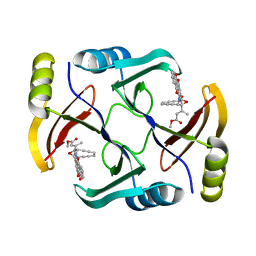 | | Crystal structure of TnmS3 in complex with tiancimycin (TNM B) | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2E)-3-[(1aS,11S,11aS,14Z,18R)-3,18-dihydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]but-2-enoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-06-05 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TnmS3 in complex with tiancimycin (TNM B)
To Be Published
|
|
6BBX
 
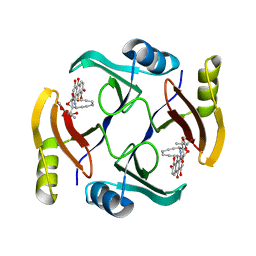 | | Crystal structure of TnmS3 in complex with TNM C | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2R,3R)-2,3-dihydroxy-3-[(1aS,11S,11aR,14Z,18R)-3,7,8,18-tetrahydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]butanoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-19 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
6CKY
 
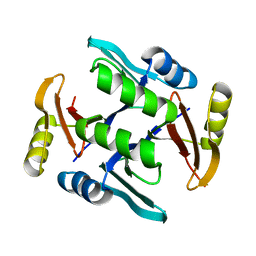 | | Crystal structure of UcmS2 | | Descriptor: | Glyoxalase | | Authors: | Chang, C.Y, Chang, C, Annaval, T, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UcmS2
To Be Published
|
|
7YIZ
 
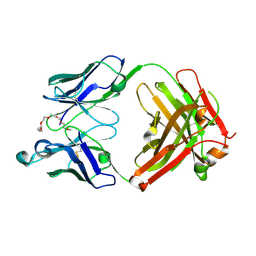 | | Crystal structure of anti-mPEG h15-2b Fab W104Y mutant | | Descriptor: | 15-2b W104Y heavy chain, 15-2b light chain, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL | | Authors: | Chang, C.Y, Nguyen, T.M.T, Li, Y.C, Su, Y.C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of anti-mPEG h15-2b Fab W104Y mutant
To Be Published
|
|
8IF7
 
 | | Crystal structure of CmnB | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CmnB | | Authors: | Chang, C.Y, Toh, S.I, Lo, C.L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of CmnB involved in the biosynthesis of the nonproteinogenic amino acid L-2,3-diaminopropionic acid.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8I80
 
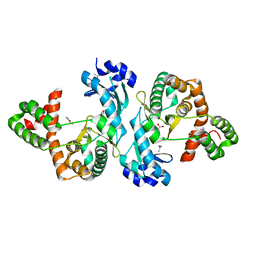 | | Crystal structure of Cph001-D189N | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J. | | Deposit date: | 2023-02-02 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I86
 
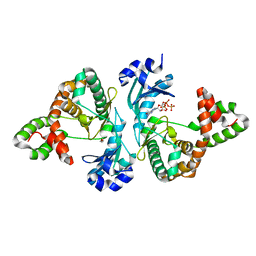 | | Crystal structure of Cph001-D189N in complex with GTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-TRIPHOSPHATE, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I84
 
 | | Crystal structure of Cph001-D189N in complex with CMN IIB | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, KBE-DPP-UAL-MYN-DPP-ALA, ... | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I85
 
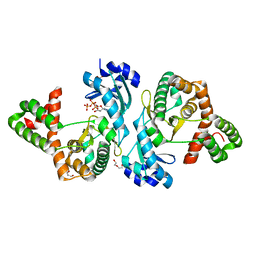 | | Crystal structure of Cph001-D189N in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I89
 
 | | Crystal structure of Cph001-D189N in complex with VIO | | Descriptor: | DI(HYDROXYETHYL)ETHER, KBE-DPP-SER-SER-UAL-5OH, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I82
 
 | |
8I8G
 
 | | Crystal structure of Cph001-D189N in complex with CMN IIA and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, KBE-DPP-UAL-MYN-DPP-SER, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I8H
 
 | | Crystal structure of Cph001-D189N in complex with VIO and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, KBE-DPP-SER-SER-UAL-5OH, ... | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
7XF9
 
 | | Crystal structure of human bleomycin hydrolase H372A mutant | | Descriptor: | Bleomycin hydrolase | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
7V5S
 
 | | Crystal structure of human bleomycin hydrolase C73A mutant | | Descriptor: | Bleomycin hydrolase, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
7V5L
 
 | | Crystal structure of human bleomycin hydrolase | | Descriptor: | Bleomycin hydrolase | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
