7EV9
 
 | | cryoEM structure of particulate methane monooxygenase associated with Cu(I) | | Descriptor: | Ammonia monooxygenase/methane monooxygenase, subunit C family protein, COPPER (I) ION, ... | | Authors: | Chang, W.H, Lin, H.H, Tsai, I.K, Huang, S.H, Chung, S.C, Tu, I.P, Yu, S.F, Chan, S.I. | | Deposit date: | 2021-05-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Copper Centers in the Cryo-EM Structure of Particulate Methane Monooxygenase Reveal the Catalytic Machinery of Methane Oxidation.
J.Am.Chem.Soc., 143, 2021
|
|
4DDN
 
 | | Structure analysis of a wound-inducible lectin ipomoelin from sweet potato | | Descriptor: | Ipomoelin, methyl alpha-D-galactopyranoside | | Authors: | Chang, W.C, Liu, K.L, Jeng, S.T, Hsu, F.C, Cheng, Y.S. | | Deposit date: | 2012-01-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ipomoelin, a jacalin-related lectin with a compact tetrameric association and versatile carbohydrate binding properties regulated by its N terminus.
Plos One, 7, 2012
|
|
3QRI
 
 | | The crystal structure of human abl1 kinase domain in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, SODIUM ION, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
3QRJ
 
 | | The crystal structure of human abl1 kinase domain T315I mutant in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
3QRK
 
 | | The crystal structure of human abl1 kinase domain in complex with DP-987 | | Descriptor: | (3S)-6-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
4C49
 
 | | Reactive loop cleaved human CBG in complex with cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, CORTICOSTEROID-BINDING GLOBULIN | | Authors: | Chan, W.L, Zhou, A, Read, R.J. | | Deposit date: | 2013-09-02 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Role of Sa-Hd Loop Movement in Cortisol Release Mechanism of Cbg
To be Published
|
|
4C41
 
 | |
3RE1
 
 | |
3BXV
 
 | |
6JJC
 
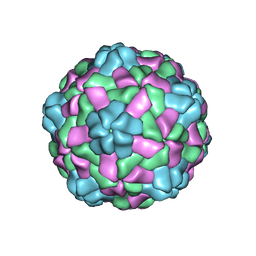 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP
To Be Published
|
|
6JJD
 
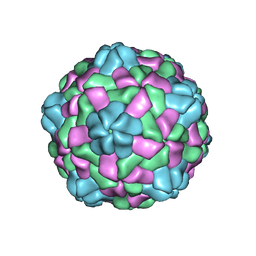 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) full VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) full VLP
To Be Published
|
|
6JJA
 
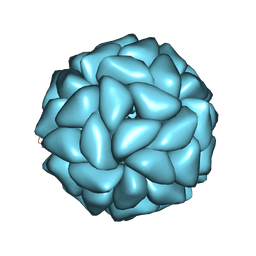 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP | | Descriptor: | CALCIUM ION, Nucleocapsid protein CP17 | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP
To Be Published
|
|
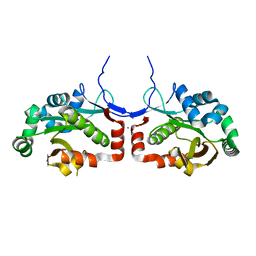
7C4H
 
 | |
1M9U
 
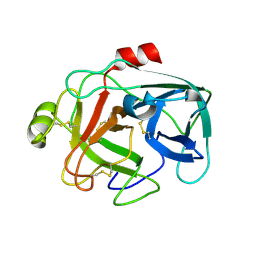 | |
1JYB
 
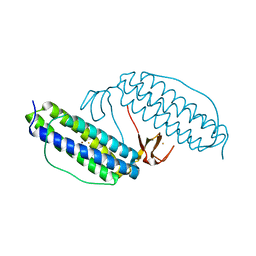 | | Crystal structure of Rubrerythrin | | Descriptor: | FE (III) ION, Rubrerythrin, ZINC ION | | Authors: | Chang, W.R, Li, M, Liu, M.Y. | | Deposit date: | 2001-09-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure studies on rubrerythrin: enzymatic activity in relation to the zinc movement.
J.Biol.Inorg.Chem., 8, 2003
|
|
2NSF
 
 | | Crystal structure of the mycothiol-dependent maleylpyruvate isomerase | | Descriptor: | GLYCEROL, Hypothetical protein Cgl3021, SULFATE ION, ... | | Authors: | Chang, W.R, Wang, R. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures and Site-directed Mutagenesis of a Mycothiol-dependent Enzyme Reveal a Novel Folding and Molecular Basis for Mycothiol-mediated Maleylpyruvate Isomerization
J.Biol.Chem., 282, 2007
|
|
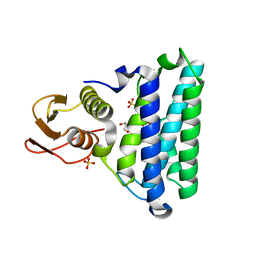
2NSG
 
 | |
4XBU
 
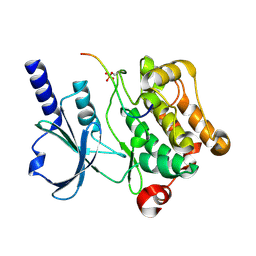 | | In vitro Crystal Structure of PAK4 in complex with Inka peptide | | Descriptor: | Protein FAM212A, Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4M6A
 
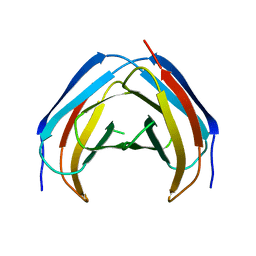 | | N-Terminal beta-Strand Swapping in a Consensus Derived Alternative Scaffold Driven by Stabilizing Hydrophobic Interactions | | Descriptor: | Tencon | | Authors: | Luo, J, Teplyakov, A, Obmolova, G, Malia, T.J, Chan, W, Jocobs, S.A, O'neil, K.T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | N-terminal beta-strand swapping in a consensus-derived alternative scaffold driven by stabilizing hydrophobic interactions.
Proteins, 82, 2014
|
|
1LN1
 
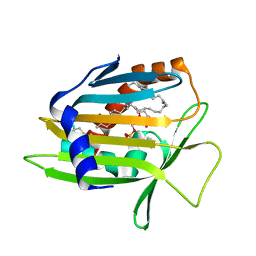 | | Crystal Structure of Human Phosphatidylcholine Transfer Protein in Complex with Dilinoleoylphosphatidylcholine | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
1LN2
 
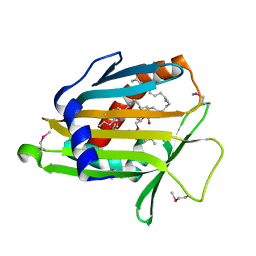 | | Crystal Structure of Human Phosphatidylcholine Transfer Protein in Complex with Dilinoleoylphosphatidylcholine (Seleno-Met Protein) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
7E37
 
 | | Crystal structure of deoxypodophyllotoxin synthase from Sinopodophyllum hexandrum in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Deoxypodophyllotoxin synthase, FE (III) ION | | Authors: | Wu, M.-H, Lin, H.-Y, Chang, W.-c, Chien, T.-C, Chan, N.-L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic analysis of carbon-carbon bond formation by deoxypodophyllotoxin synthase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E38
 
 | | Crystal structure of deoxypodophyllotoxin synthase from Sinopodophyllum hexandrum in complex with yatein and succinate | | Descriptor: | (3~{R},4~{R})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, (3~{S},4~{S})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, Deoxypodophyllotoxin synthase, ... | | Authors: | Wu, M.-H, Chang, W.-c, Chien, T.-C, Chan, N.-L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic analysis of carbon-carbon bond formation by deoxypodophyllotoxin synthase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3JCU
 
 | | Cryo-EM structure of spinach PSII-LHCII supercomplex at 3.2 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wei, X.P, Zhang, X.Z, Su, X.D, Cao, P, Liu, X.Y, Li, M, Chang, W.R, Liu, Z.F. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of spinach photosystem II-LHCII supercomplex at 3.2 A resolution
Nature, 534, 2016
|
|
3JB6
 
 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA-dependent RNA polymerase, VP1 CSP, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-02 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
