9IQO
 
 | |
3J9B
 
 | | Electron cryo-microscopy of an RNA polymerase | | Descriptor: | Polymerase, Polymerase basic protein 2, RNA (5'-R(*UP*UP*UP*UP*UP*A)-3'), ... | | Authors: | Chang, S.H, Sun, D.P, Liang, H.H, Wang, J, Li, J, Guo, L, Wang, X.L, Guan, C.C, Boruah, B.M, Yuan, L.M, Feng, F, Yang, M.R, Wojdyla, J, Wang, J.W, Wang, M.T, Wang, H.W, Liu, Y.F. | | Deposit date: | 2014-12-16 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3 angstrom Resolution.
Mol.Cell, 2015
|
|
6KAF
 
 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1W42
 
 | | T. celer L30e R92A variant | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Lee, C.F, Lee, K.M, Chan, S.H, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-22 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Electrostatic Interactions Contribute to Reduced Heat Capacity Change of Unfolding in a Thermophilic Ribosomal Protein L30E
J.Mol.Biol., 348, 2005
|
|
1W40
 
 | | T. celer L30e K9A variant | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Lee, C.F, Lee, K.M, Chan, S.H, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-22 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Electrostatic Interactions Contribute to Reduced Heat Capacity Change of Unfolding in a Thermophilic Ribosomal Protein L30E
J.Mol.Biol., 348, 2005
|
|
1W41
 
 | | T. celer L30e E90A variant | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Lee, C.F, Lee, K.M, Chan, S.H, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-22 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Electrostatic Interactions Contribute to Reduced Heat Capacity Change of Unfolding in a Thermophilic Ribosomal Protein L30E
J.Mol.Biol., 348, 2005
|
|
6G4A
 
 | | FLN5 (full length) | | Descriptor: | Gelation factor | | Authors: | Waudby, C.A, Wlodarski, T, Karyadi, M.-E, Cassaignau, A.M.E, Chan, S.H.S, Wentink, A.S, Schmidt-Engler, J.M, Camilloni, C, Vendruscolo, M, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mapping energy landscapes of a growing filamin domain reveals an intermediate associated with proline isomerization during biosynthesis
To Be Published
|
|
4CAU
 
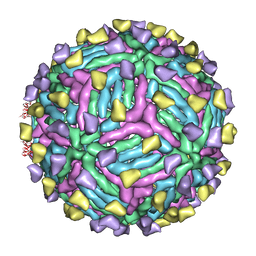 | | THREE-DIMENSIONAL STRUCTURE OF DENGUE VIRUS SEROTYPE 1 COMPLEXED WITH 2 HMAB 14C10 FAB | | Descriptor: | ENVELOPE PROTEIN E, FAB 14C10 | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A.P, Tan, T.T, Yip, A, Schul, W, Aung, M, Kostyuchenko, V.A, Leo, Y.S, Chan, S.H, Smith, K.G, Chan, A.H, Zou, G, Ooi, E.E, Kemeny, D.M, Tan, G.K, Ng, J.K, Ng, M.L, Alonso, S, Fisher, D, Shi, P.Y, Hanson, B.J, Lok, S.M, Macary, P.A. | | Deposit date: | 2013-10-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The Structural Basis for Serotype-Specific Neutralization of Dengue Virus by a Human Antibody.
Sci.Trans.Med, 4, 2012
|
|
7ZP8
 
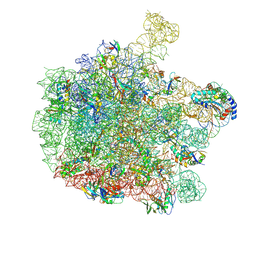 | | 70S E. coli ribosome with a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Chan, S.H.S, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
4ZCF
 
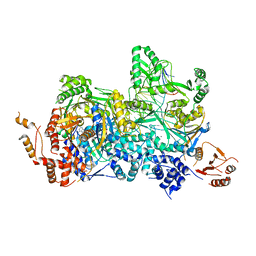 | | Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, DNA 20-mer AATCATAGTCTACTGCTGTA, ... | | Authors: | Gupta, Y.K, Chan, S.H, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I.
Nat Commun, 6, 2015
|
|
3J05
 
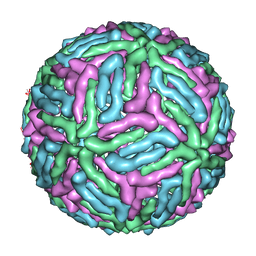 | | Three-dimensional structure of Dengue virus serotype 1 complexed with HMAb 14c10 Fab | | Descriptor: | envelope protein | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A, Tan, T.T, Shi, P.Y, Yip, A, Schul, W, Leo, Y.S, Chan, S.H, Smith, K.G.C, Ooi, E.E, Kemeny, D.M, Ng, G, Ng, M.L, Alonso, S, Fisher, D, Hanson, B, Lok, S.M, MacAry, P.A. | | Deposit date: | 2011-04-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The structural basis for serotype-specific neutralization of dengue virus by a human antibody.
Sci Transl Med, 4, 2012
|
|
2B5A
 
 | | C.BclI, Control Element of the BclI Restriction-Modification System | | Descriptor: | ACETIC ACID, C.BclI | | Authors: | Sawaya, M.R, Zhu, Z, Mersha, F, Chan, S.H, Dabur, R, Xu, S.Y, Balendiran, G.K. | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Crystal Structure of the Restriction-Modification System Control Element C.BclI and Mapping of Its Binding Site.
Structure, 13, 2005
|
|
3O8L
 
 | | Structure of phosphofructokinase from rabbit skeletal muscle | | Descriptor: | 6-phosphofructokinase, muscle type, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Banaszak, K, Chang, S.H, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
3O8N
 
 | | Structure of phosphofructokinase from rabbit skeletal muscle | | Descriptor: | 6-phosphofructokinase, muscle type, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Banaszak, K, Chang, S.H, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
1ZD0
 
 | | Crystal structure of Pfu-542154 conserved hypothetical protein | | Descriptor: | MAGNESIUM ION, METHANOL, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J.E, Liu, Z.J, Horanyi, P.S, Florence, Q.J.T, Tempel, W, Zhou, W, Chen, L, Lee, D, Nguyen, J, Chang, S.H, Bereton, P, Izumi, M, Jenny Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-13 | | Release date: | 2005-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Pfu-542154 conserved hypothetical protein
To be Published
|
|
6S8G
 
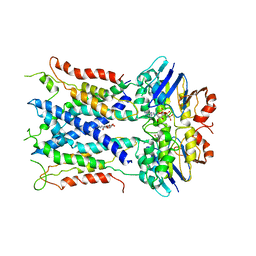 | | Cryo-EM structure of LptB2FGC in complex with AMP-PNP | | Descriptor: | Inner membrane protein yjgQ, LPS export ABC transporter permease LptF, Lipopolysaccharide ABC transporter, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8N
 
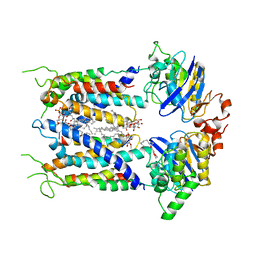 | | Cryo-EM structure of LptB2FGC in complex with lipopolysaccharide | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, Inner membrane protein yjgQ, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8H
 
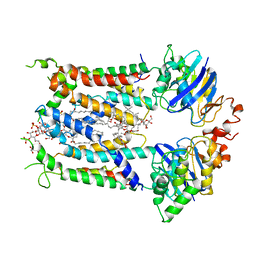 | | Cryo-EM structure of LptB2FG in complex with LPS | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
7E80
 
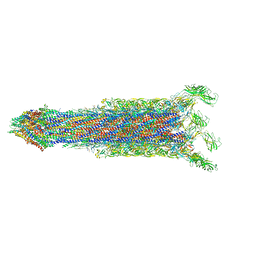 | | Cryo-EM structure of the flagellar rod with hook and export apparatus from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E81
 
 | | Cryo-EM structure of the flagellar MS ring with FlgB-Dc loop and FliE-helix 1 from Salmonella | | Descriptor: | Flagellar M-ring protein, FlgB-Dc loop, FliE helix 1 | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E82
 
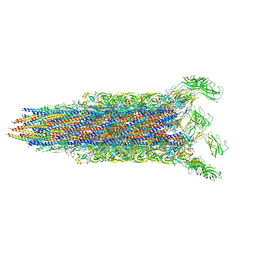 | | Cryo-EM structure of the flagellar rod with partial hook from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7ARM
 
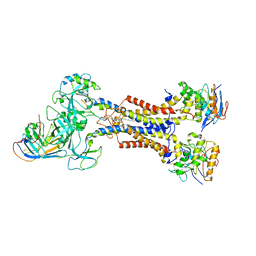 | | LolCDE in complex with lipoprotein and LolA | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARL
 
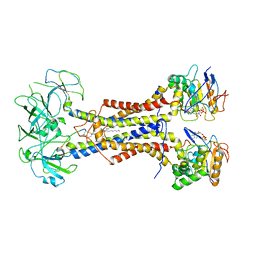 | | LolCDE in complex with lipoprotein and ADP | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, ADENOSINE-5'-DIPHOSPHATE, LPP, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARK
 
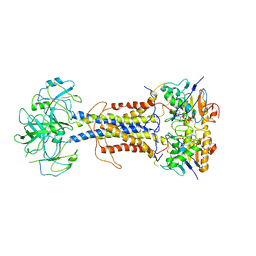 | | LolCDE in complex with AMP-PNP in the closed NBD state | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARI
 
 | | LolCDE apo structure | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
