4E4G
 
 | | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
4E6M
 
 | | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium)
To be Published
|
|
4HAD
 
 | | Crystal structure of probable oxidoreductase protein from Rhizobium etli CFN 42 | | Descriptor: | Probable oxidoreductase protein, SODIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-09-26 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of probable oxidoreductase protein from Rhizobium etli CFN 42
To be Published
|
|
4E3E
 
 | | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl | | Descriptor: | MaoC domain protein dehydratase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl
To be Published
|
|
4E3A
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 | | Descriptor: | ADENOSINE, sugar kinase protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42
To be Published
|
|
4GWB
 
 | | Crystal structure of putative Peptide methionine sulfoxide reductase from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Peptide methionine sulfoxide reductase MsrA 3 | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-09-01 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative Peptide methionine sulfoxide reductase from Sinorhizobium meliloti 1021
To be Published
|
|
1XEO
 
 | | High Resolution Crystals Structure of Cobalt- Peptide Deformylase Bound To Formate | | Descriptor: | COBALT (II) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
1XEM
 
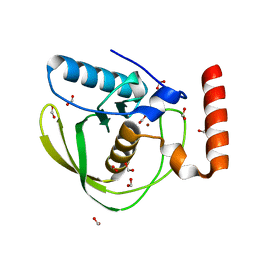 | | High Resolution Crystal Structure of Escherichia coli Zinc- Peptide Deformylase bound to formate | | Descriptor: | FORMIC ACID, Peptide deformylase, ZINC ION | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
1XEN
 
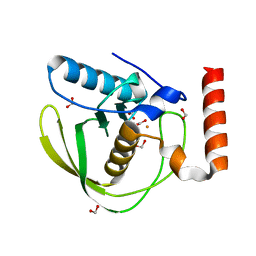 | | High Resolution Crystal Structure of Escherichia coli Iron- Peptide Deformylase Bound To Formate | | Descriptor: | FE (III) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
1TUE
 
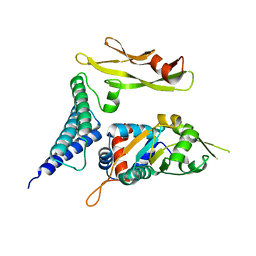 | |
4J3Z
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Jannaschia sp. CCS1 | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Jannaschia sp. CCS1
To be Published
|
|
4IX1
 
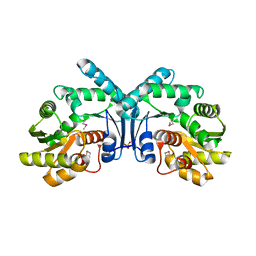 | | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205 | | Descriptor: | PHOSPHATE ION, hypothetical protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205
To be Published
|
|
4K9C
 
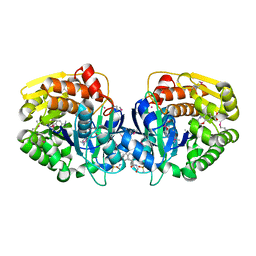 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE and 4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-CARBOXYLIC ACID | | Descriptor: | 4-methyl-3,4-dihydro-2H-1,4-benzoxazine-7-carboxylic acid, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE and 4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-CARBOXYLIC ACID
To be Published
|
|
4IYM
 
 | | Crystal structure of putative methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD, target 011934 | | Descriptor: | MAGNESIUM ION, Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-28 | | Release date: | 2013-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD, target 011934
To be Published
|
|
4J6F
 
 | | Crystal structure of putative alcohol dehydrogenase from Sinorhizobium meliloti 1021, NYSGRC-Target 012230 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative alcohol dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-11 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative alcohol dehydrogenase from Sinorhizobium meliloti 1021, NYSGRC-Target 012230
To be Published
|
|
4JHS
 
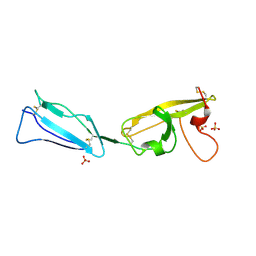 | | Crystal structure of a C-terminal two domain fragment of human beta-2-glycoprotein 1 | | Descriptor: | Beta-2-glycoprotein 1, SULFATE ION | | Authors: | Bonanno, J.B, Toro, R, Gizzi, A, Chan, M.K, Garrett-Thomson, S.C, Patel, H, Lim, S, Matikainen, B, Celikgil, A, Garforth, S, Hillerich, B, Seidel, R, Rand, J.H, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a C-terminal two domain fragment of human beta-2-glycoprotein 1
To be Published
|
|
4K8C
 
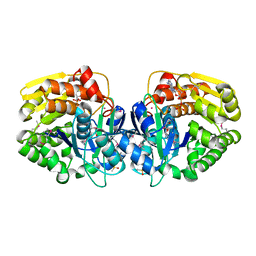 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ADP | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ADP
To be Published
|
|
4K93
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, N-(hydroxymethyl)benzamide, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE
To be Published
|
|
4K8T
 
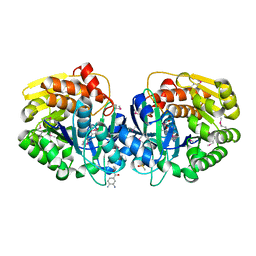 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ethyl 3,4-diaminobenzoate | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ethyl 3,4-diaminobenzoate
To be Published
|
|
4KBE
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with benzoguanamine | | Descriptor: | 6-phenyl-1,3,5-triazine-2,4-diamine, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with benzoguanamine
To be Published
|
|
4K8P
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-ethylbenzyl alcohol | | Descriptor: | (2-ETHYLPHENYL)METHANOL, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-ethylbenzyl alcohol
To be Published
|
|
4K9Z
 
 | | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P6222 | | Descriptor: | CHLORIDE ION, Putative thiol-disulfide oxidoreductase | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R.D, Bonanno, J.B, Armstrong, R.N, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P6222
To be Published
|
|
4KAN
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid | | Descriptor: | (2,5-dimethyl-1,3-thiazol-4-yl)acetic acid, ADENOSINE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid
To be Published
|
|
4KAD
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N1-(2.3-dihydro-1H-inden-5-yl)acetam | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, N-(4,7-dihydro-1H-inden-6-yl)acetamide, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N1-(2.3-dihydro-1H-inden-5-yl)acetam
To be Published
|
|
4K9I
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with Norharmane | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, Norharmane, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with Norharmane
To be Published
|
|
