7TBH
 
 | | cryo-EM structure of MBP-KIX-apoferritin complex with peptide 7 | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed, LEU-SER-ARG-ARG-PRO-SEP-TYR-ARG-LYS-ILE-LEU-ASN-ASP-LEU-SER-SER-ASP-ALA-PRO | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
7TB3
 
 | | cryo-EM structure of MBP-KIX-apoferritin | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
5NJT
 
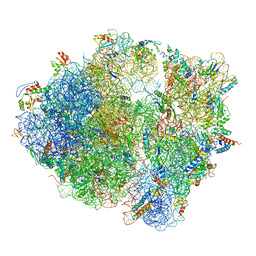 | | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Beckert, B, Abdelshahid, M, Schaefer, H, Steinchen, W, Arenz, S, Berninghausen, O, Beckmann, R, Bange, G, Turgay, K, Wilson, D.N. | | Deposit date: | 2017-03-29 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization.
EMBO J., 36, 2017
|
|
2I1B
 
 | |
3WIR
 
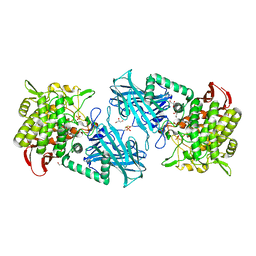 | | Crystal structure of kojibiose phosphorylase complexed with glucose | | Descriptor: | GLYCEROL, Kojibiose phosphorylase, PHOSPHATE ION, ... | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
3WIQ
 
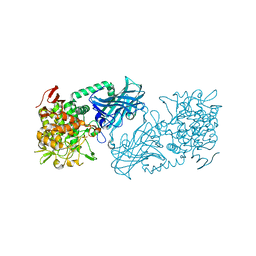 | | Crystal structure of kojibiose phosphorylase complexed with kojibiose | | Descriptor: | Kojibiose phosphorylase, SULFATE ION, alpha-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
6SHR
 
 | | X-RAY CRYSTAL STRUCTURE OF CELL-FREE PROTEIN SYNTHESIS (CFPS) PRODUCED SDF1-A | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Jugnarain, V.M, Mitchell, E, Forsyth, T, Michael, H, Cortes, S, Tillier, B. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | X-RAY CRYSTAL STRUCTURE OF CELL-FREE PROTEIN SYNTHESIS (CFPS) PRODUCED SDF1-A
To Be Published
|
|
