8JYL
 
 | | Acyl-ACP Synthetase structure bound to C10-AMS | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-decanoylsulfamate | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Acyl-ACP Synthetase structure bound to C10-AMS
To Be Published
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
8IL2
 
 | |
8IL6
 
 | |
4JJH
 
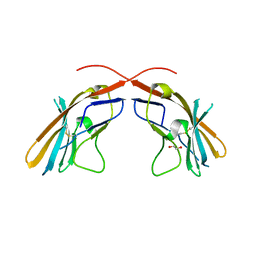 | | Crystal structure of the D1 domain from human Nectin-4 extracellular fragment [PSI-NYSGRC-005624] | | Descriptor: | 1,2-ETHANEDIOL, Poliovirus receptor-related protein 4 | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D1 domain of human Nectin-4 extracellular fragment [NYSGRC-005624]
to be published
|
|
8CYE
 
 | |
8CVI
 
 | |
8IKQ
 
 | |
8IL1
 
 | |
6KKR
 
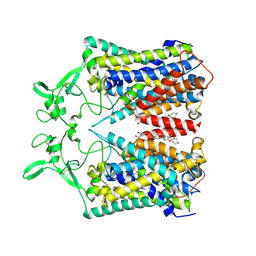 | | human KCC1 structure determined in KCl and detergent GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
6KKT
 
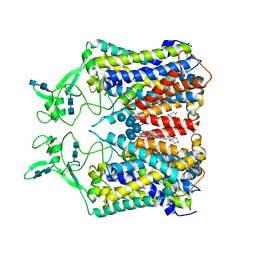 | | human KCC1 structure determined in KCl and lipid nanodisc | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
7OFG
 
 | |
4JGJ
 
 | | Crystal structure of the Ig-like D1 domain from mouse Carcinoembryogenic antigen-related cell adhesion molecule 15 (CEACAM15) [PSI-NYSGRC-005691] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 15, Unknown peptide | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6508 Å) | | Cite: | Crystal structure of the Ig-like D1 domain of CEACAM15 from Mus musculus [NYSGRC-005691]
to be published
|
|
8RQ9
 
 | | Crystal structure of PROTAC CFT-1297 in complex with CRBN-midi and BRD4(BD2) | | Descriptor: | (3~{S})-3-[7-[1-[7-[4-[6-(4-chlorophenyl)-1-methyl-spiro[[1,2,4]triazolo[4,3-a][1,4]benzodiazepine-4,1'-cyclopropane]-8-yl]pyrazol-1-yl]heptyl]piperidin-4-yl]-3-oxidanylidene-1~{H}-isoindol-2-yl]piperidine-2,6-dione, Bromodomain-containing protein 4, Protein cereblon, ... | | Authors: | Darren, D, Ramachandran, S, Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
6KKU
 
 | | human KCC1 structure determined in NaCl and GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
4ZXF
 
 | | Crystal Structure of a Soluble Variant of Monoglyceride Lipase from Saccharomyces Cerevisiae in Complex with a Substrate Analog | | Descriptor: | 1-{3-[(R)-hydroxy(octadecyloxy)phosphoryl]propyl}triaza-1,2-dien-2-ium, Monoglyceride lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Lichtenegger, J, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
3GG2
 
 | | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate | | Descriptor: | Sugar dehydrogenase, UDP-glucose/GDP-mannose dehydrogenase family, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate
To be Published
|
|
8R0S
 
 | | Structure of reverse transcriptase from Cauliflower Mosaic Virus in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*CP*GP*CP*AP*CP*TP*GP*CP*TP*GP*GP*A)-3'), Enzymatic polyprotein, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*AP*GP*UP*GP*CP*GP*UP*AP*GP*C)-3') | | Authors: | Prabaharan, C, Figiel, M, Chamera, S, Szczepanowski, R, Nowak, E, Nowotny, M. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical characterization of cauliflower mosaic virus reverse transcriptase.
J.Biol.Chem., 300, 2024
|
|
5XR1
 
 | |
3GKB
 
 | | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis
To be Published
|
|
8QN4
 
 | | Structure of BAM-EspP complex in the non-closing EspP state | | Descriptor: | EspP epsilon, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Xie, T, Pang, J, Shen, C, Chang, S, Tang, X, Zhang, X, Dong, H, Zhou, R. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Dynamic topology-mediated maturation of beta-barrel proteins in BAM-catalyzed folding
To Be Published
|
|
5N2M
 
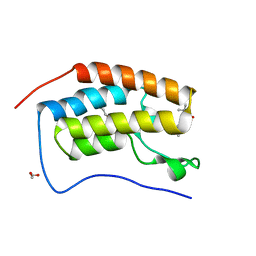 | | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, propan-2-yl ~{N}-[(2~{S},4~{R})-6-(3-acetamidophenyl)-1-ethanoyl-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
3ER6
 
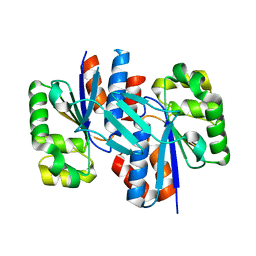 | | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus | | Descriptor: | Putative transcriptional regulator protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus
To be Published
|
|
7NBU
 
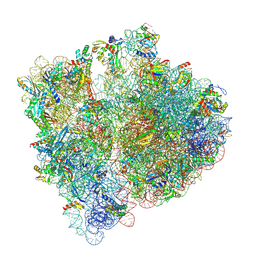 | | Structure of the HigB1 toxin mutant K95A from Mycobacterium tuberculosis (Rv1955) and its target, the cspA mRNA, on the E. coli Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Giudice, E, Mansour, M, Chat, S, D'Urso, G, Gillet, R, Genevaux, P. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Substrate recognition and cryo-EM structure of the ribosome-bound TAC toxin of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
