4KAH
 
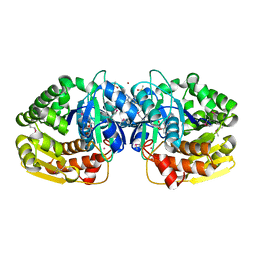 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 4-bromo-1H-pyrazole | | Descriptor: | 4-bromo-1H-pyrazole, ADENOSINE, BROMIDE ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 4-bromo-1H-pyrazole
To be Published
|
|
4K9I
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with Norharmane | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, Norharmane, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with Norharmane
To be Published
|
|
4KA0
 
 | | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P21221 | | Descriptor: | CHLORIDE ION, Putative thiol-disulfide oxidoreductase | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R.D, Bonanno, J.B, Armstrong, R.N, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-21 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P21221
To be Published
|
|
4M1E
 
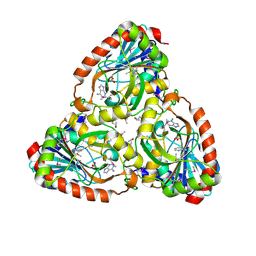 | | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364. | | Descriptor: | ADENINE, PYRIDINE-2-CARBOXYLIC ACID, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364.
To be Published
|
|
4MAR
 
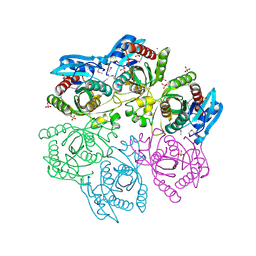 | | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279 complexed with sulfate. | | Descriptor: | MAGNESIUM ION, Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-16 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279 complexed with sulfate.
To be Published
|
|
4M3N
 
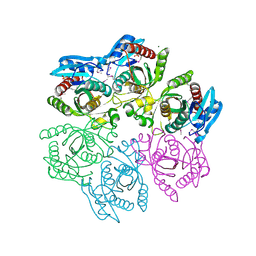 | | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279, NYSGRC Target 029804. | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-06 | | Release date: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279, NYSGRC Target 029804.
To be Published
|
|
4LZA
 
 | | Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700. | | Descriptor: | Adenine phosphoribosyltransferase, CHLORIDE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700.
To be Published
|
|
4MCH
 
 | | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520. | | Descriptor: | 6-hydroxynaphthalene-1-carboxylic acid, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520.
To be Published
|
|
4M0K
 
 | | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CALCIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775.
To be Published
|
|
4M1Q
 
 | | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, L-lactate dehydrogenase, PHOSPHATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-03 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814.
To be Published
|
|
4M7W
 
 | | Crystal structure of purine nucleoside phosphorylase from Leptotrichia buccalis C-1013-b, NYSGRC Target 029767. | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-12 | | Release date: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Leptotrichia buccalis C-1013-b, NYSGRC Target 029767.
To be Published
|
|
7XXN
 
 | | HapR Quadruple mutant, bound to Qstatin | | Descriptor: | 1-(5-bromanylthiophen-2-yl)sulfonylpyrazole, GLYCEROL, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY0
 
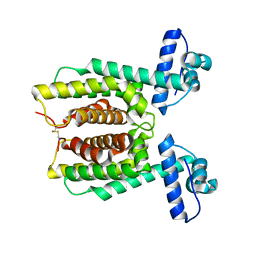 | | HapR Double mutant Y76F, F171C | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7Y4J
 
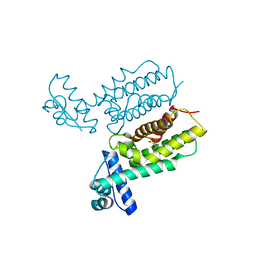 | | HapR_Triple mutant Y76F, L97I, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXS
 
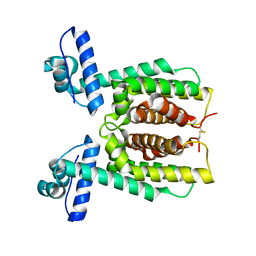 | | HapR mutant I141V | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XYI
 
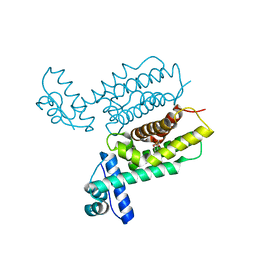 | | HapR Quadruple mutant Y76F, L97I, I141V, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXO
 
 | | HapR Native in CHES buffer pH 9.5 | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY5
 
 | | HapR_Double Mutant with CHES buffer | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXT
 
 | | HapR Quadruple mutant, bound to IMTVC-212 | | Descriptor: | 1-((5-phenylthiophen-2-yl)sulfonyl)-1H-pyrazole, DIMETHYL SULFOXIDE, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
6X46
 
 | | NMR solution structure of Asterix/Gtsf1 from mouse (CHHC zinc finger domains) | | Descriptor: | Gametocyte-specific factor 1, ZINC ION | | Authors: | Ipsaro, J.J, O'Brien, P.A, Bhattacharya, S, Palmer III, A.G, Joshua-Tor, L. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Asterix/Gtsf1 links tRNAs and piRNA silencing of retrotransposons.
Cell Rep, 34, 2021
|
|
8H5V
 
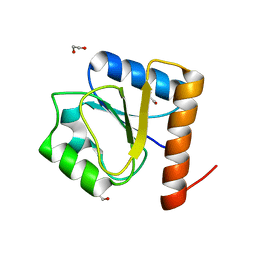 | | Crystal structure of the FleQ domain of Vibrio cholerae FlrA | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, flagellar regulatory protein A | | Authors: | Dasgupta, J, Chakraborty, S. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The N-terminal FleQ domain of the Vibrio cholerae flagellar master regulator FlrA plays pivotal structural roles in stabilizing its active state.
Febs Lett., 597, 2023
|
|
7M6M
 
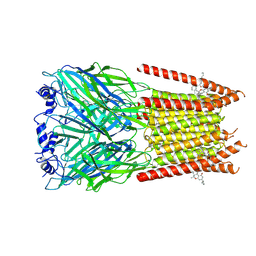 | | Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6N
 
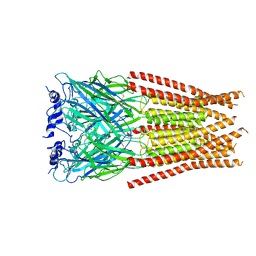 | |
7M6Q
 
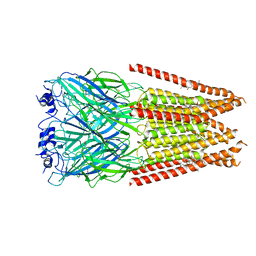 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1 | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6R
 
 | |
