1P50
 
 | | Transition state structure of an Arginine Kinase mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
3RVT
 
 | | Structure of 4C1 Fab in P212121 space group | | Descriptor: | Fab fragment of 4C1 antibody - heavy chain, Fab fragment of 4C1 antibody - light chain | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular determinants for antibody binding on group 1 house dust mite allergens.
J.Biol.Chem., 287, 2012
|
|
3UFY
 
 | | AKR1C3 complex with R-naproxen | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, ... | | Authors: | Squire, C.J, Flanagan, J.U, Yosaatmadja, Y, Teague, R.M, Chai, M. | | Deposit date: | 2011-11-01 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
3E22
 
 | | Tubulin-colchicine-soblidotin: Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cormier, A, Marchand, M, Ravelli, R.B, Knossow, M, Gigant, B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insight into the inhibition of tubulin by vinca domain peptide ligands
Embo Rep., 9, 2008
|
|
1P52
 
 | | Structure of Arginine kinase E314D mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, D-ARGININE, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
1OMI
 
 | | Crystal structure of PrfA,the transcriptional regulator in Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeriolysin regulatory protein | | Authors: | Thirumuruhan, R, Rajashankar, K, Fedorov, A.A, Dodatko, T, Chance, M.R, Cossart, P, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-02-25 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PrfA, the transcriptional regulator in Listeria monocytogenes
To be Published
|
|
3RVU
 
 | | Structure of 4C1 Fab in C2221 space group | | Descriptor: | 4C1 Fab - heavy chain, 4C1 Fab - light chain | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular determinants for antibody binding on group 1 house dust mite allergens.
J.Biol.Chem., 287, 2012
|
|
4HU7
 
 | | E. coli thioredoxin variant with Pro76 as single proline residue | | Descriptor: | COPPER (II) ION, SODIUM ION, Thioredoxin-1 | | Authors: | Glockshuber, R, Scharer, M.A, Capitani, G, Rubini, M. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | (4R)- and (4S)-Fluoroproline in the Conserved cis-Prolyl Peptide Bond of the Thioredoxin Fold: Tertiary Structure Context Dictates Ring Puckering.
Chembiochem, 14, 2013
|
|
3TVI
 
 | | Crystal structure of Clostridium acetobutylicum aspartate kinase (CaAK): An important allosteric enzyme for industrial amino acids production | | Descriptor: | ASPARTIC ACID, Aspartokinase, LYSINE | | Authors: | Manjasetty, B.A, Chance, M.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-09-20 | | Release date: | 2011-11-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Clostridium acetobutylicum Aspartate kinase (CaAK): An important allosteric enzyme for amino acids production.
Biotechnol Rep (Amst), 3, 2014
|
|
3J4P
 
 | | Electron Microscopy Analysis of a Disaccharide Analog complex Reveals Receptor Interactions of Adeno-Associated Virus | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Capsid protein VP1, MAGNESIUM ION, ... | | Authors: | Xie, Q, Chapman, M.S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Electron microscopy analysis of a disaccharide analog complex reveals receptor interactions of adeno-associated virus.
J.Struct.Biol., 184, 2013
|
|
3TQ7
 
 | | EB1c/EB3c heterodimer in complex with the CAP-Gly domain of P150glued | | Descriptor: | Dynactin subunit 1, Microtubule-associated protein RP/EB family member 1, Microtubule-associated protein RP/EB family member 3 | | Authors: | De Groot, C.O, Scharer, M.A, Capitani, G, Steinmetz, M.O. | | Deposit date: | 2011-09-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interaction of mammalian end binding proteins with CAP-Gly domains of CLIP-170 and p150(glued).
J.Struct.Biol., 177, 2012
|
|
3J4A
 
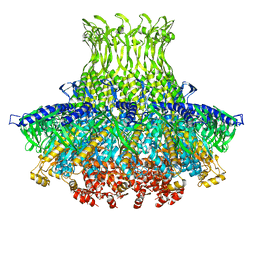 | | Structure of gp8 connector protein | | Descriptor: | Head-to-tail joining protein | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
1CZP
 
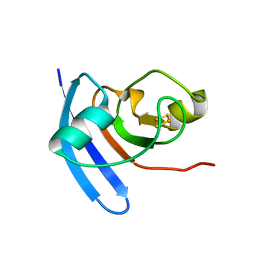 | | ANABAENA PCC7119 [2FE-2S] FERREDOXIN IN THE REDUCED AND OXIXIZED STATE AT 1.17 A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Morales, R, Charon, M.H, Frey, M. | | Deposit date: | 1999-09-06 | | Release date: | 2000-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Refined X-ray structures of the oxidized, at 1.3 A, and reduced, at 1.17 A, [2Fe-2S] ferredoxin from the cyanobacterium Anabaena PCC7119 show redox-linked conformational changes.
Biochemistry, 38, 1999
|
|
1PRB
 
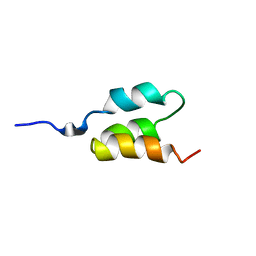 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN PAB | | Authors: | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
1QVX
 
 | | SOLUTION STRUCTURE OF THE FAT DOMAIN OF FOCAL ADHESION KINASE | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Gao, G, Prutzman, K.C, King, M.L, DeRose, E.F, London, R.E, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Focal Adhesion Targeting Domain of Focal Adhesion Kinase in Complex with a Paxillin LD Peptide: EVIDENCE FOR A TWO-SITE BINDING MODEL.
J.Biol.Chem., 279, 2004
|
|
1QQH
 
 | |
4HS3
 
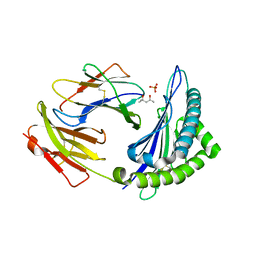 | | Crystal structure of H-2Kb with a disulfide stabilized F pocket in complex with the LCMV derived peptide GP34 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, Envelope glycoprotein, ... | | Authors: | Uchtenhagen, H, Boulanger, B, Hein, Z, Abualrous, E.T, Zacharias, M, Werner, J, Elliott, T, Springer, S, Achour, A. | | Deposit date: | 2012-10-29 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide-independent stabilization of MHC class I molecules breaches cellular quality control.
J.Cell.Sci., 127, 2014
|
|
1QFI
 
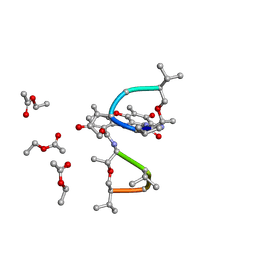 | | SYNTHESIS AND STRUCTURE OF PROLINE RING MODIFIED ACTINOMYCINS OF X TYPE | | Descriptor: | ACTINOMYCIN X2, ETHYL ACETATE, METHANOL | | Authors: | Lifferth, A, Bahner, I, Lackner, H, Schaefer, M. | | Deposit date: | 1999-04-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Synthesis and Structure of Proline Ring Modified Actinomycins of the X-Type
Z.Naturforsch., 54, 1999
|
|
2AL6
 
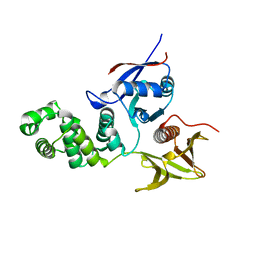 | | FERM domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
2XJ0
 
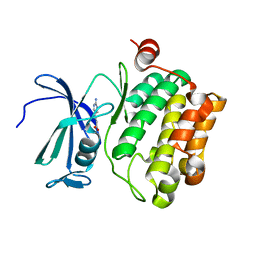 | | Protein kinase Pim-1 in complex with fragment-4 from crystallographic fragment screen | | Descriptor: | (E)-3-(2-AMINO-PYRIDINE-5YL)-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2AVK
 
 | | met-azido-DcrH-Hr | | Descriptor: | MONOAZIDO-MU-OXO-DIIRON, SULFATE ION, hemerythrin-like domain protein DcrH | | Authors: | Isaza, C.E, Silaghi-Dumitrescu, R, Iyer, R.B, Kurtz, D.M, Chan, M.K. | | Deposit date: | 2005-08-30 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Basis for O(2) Sensing by the Hemerythrin-like Domain of a Bacterial Chemotaxis Protein: Substrate Tunnel and Fluxional N Terminus.
Biochemistry, 45, 2006
|
|
2XM3
 
 | | Deinococcus radiodurans ISDra2 Transposase Left end DNA complex | | Descriptor: | 5'-D(*TP*TP*AP*GP*T)-3', ACETATE ION, DRA2 TRANSPOSASE BINDING ELEMENT, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XSE
 
 | | The structural basis for recognition of J-base containing DNA by a novel DNA-binding domain in JBP1 | | Descriptor: | GLYCEROL, NITRATE ION, THYMINE DIOXYGENASE JBP1 | | Authors: | Heidebrecht, T, Christodoulou, E, Chalmers, M.J, Jan, S, ter Riete, B, Grover, R.K, Joosten, R.P, Littler, D, vanLuenen, H, Griffin, P.R, Wentworth, P, Borst, P, Perrakis, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Recognition of Base J Containing DNA by a Novel DNA Binding Domain in Jbp1.
Nucleic Acids Res., 39, 2011
|
|
1QSZ
 
 | | THE VEGF-BINDING DOMAIN OF FLT-1 (MINIMIZED MEAN) | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 1 | | Authors: | Starovasnik, M.A, Christinger, H.W, Wiesmann, C, Champe, M.A, de Vos, A.M, Skelton, N.J. | | Deposit date: | 1999-06-24 | | Release date: | 1999-11-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the VEGF-binding domain of Flt-1: comparison of its free and bound states.
J.Mol.Biol., 293, 1999
|
|
2YGD
 
 | | Molecular architectures of the 24meric eye lens chaperone alphaB- crystallin elucidated by a triple hybrid approach | | Descriptor: | ALPHA-CRYSTALLIN B CHAIN | | Authors: | Braun, N, Zacharias, M, Peschek, J, Kastenmueller, A, Zou, J, Hanzlik, M, Haslbeck, M, Rappsilber, J, Buchner, J, Weinkauf, S. | | Deposit date: | 2011-04-13 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Multiple Molecular Architectures of the Eye Lens Chaperone Alpha Beta-Crystallin Elucidated by a Triple Hybrid Approach
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
