1XI3
 
 | | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001 | | Descriptor: | NICKEL (II) ION, SULFATE ION, Thiamine phosphate pyrophosphorylase, ... | | Authors: | Zhou, W, Chen, L, Tempel, W, Liu, Z.-J, Habel, J, Lee, D, Lin, D, Chang, S.-H, Dillard, B.D, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Kelley, L.-L.C, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001
To be published
|
|
5N88
 
 | | Crystal structure of antibody bound to viral protein | | Descriptor: | PC4 and SFRS1-interacting protein, VH59 antibody | | Authors: | Bao, L, Hannon, C, Cruz-Migoni, A, Ptchelkine, D, Sun, M.-y, Derveni, M, Bunjobpol, W, Chambers, J.S, Simmons, A, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intracellular immunization against HIV infection with an intracellular antibody that mimics HIV integrase binding to the cellular LEDGF protein.
Sci Rep, 7, 2017
|
|
5U3F
 
 | | Structure of Mycobacterium tuberculosis IlvE, a branched-chain amino acid transaminase, in complex with D-cycloserine derivative | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Branched-chain-amino-acid aminotransferase | | Authors: | Favrot, L, Amorim Franco, T.M, Blanchard, J.S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Mechanism-Based Inhibition of the Mycobacterium tuberculosis Branched-Chain Aminotransferase by d- and l-Cycloserine.
ACS Chem. Biol., 12, 2017
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
7ULN
 
 | | Turnip yellows virus N-terminal readthrough domain | | Descriptor: | Minor capsid protein P3-RTD | | Authors: | Schiltz, C.J, Chappie, J.S. | | Deposit date: | 2022-04-05 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Polerovirus N-terminal readthrough domain structures reveal molecular strategies for mitigating virus transmission by aphids
Nat Commun, 13, 2022
|
|
7ULO
 
 | |
3GUB
 
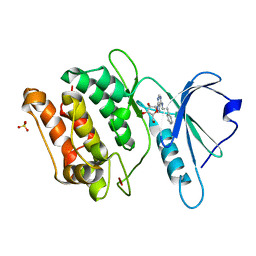 | | Crystal structure of DAPKL93G complexed with N6-(2-Phenylethyl)adenosine | | Descriptor: | 9-alpha-L-lyxofuranosyl-N-(2-phenylethyl)-9H-purin-6-amine, Death-associated protein kinase 1, SULFATE ION | | Authors: | McNamara, L.K, Schumacher, A.M, Schavocky, J.S, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2009-03-29 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structures of the DAPK gatekeeper mutant complexed with N6-modified adenosine analogs.
To be Published
|
|
3GU7
 
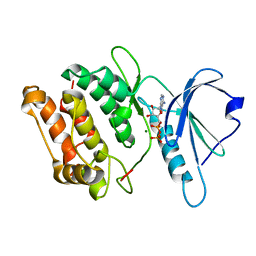 | |
3GU5
 
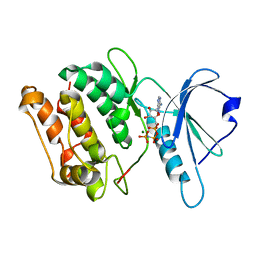 | |
3GU8
 
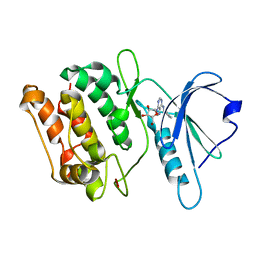 | | Crystal structure of DAPKL93G with N6-cyclopentyladenosine | | Descriptor: | Death-associated protein kinase 1, N6-cyclopentyladenosine | | Authors: | McNamara, L.K, Schumacher, A.M, Schavocky, J.S, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2009-03-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the DAPK gatekeeper mutant with N6-modified adenosine analogs.
To be Published
|
|
3GU4
 
 | |
3GU6
 
 | |
3TUC
 
 | | Crystal structure of SYK kinase domain with 1-benzyl-N-(5-(6,7-dimethoxyquinolin-4-yloxy)pyridin-2-yl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Descriptor: | 1-benzyl-N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-2-oxo-1,2-dihydropyridine-3-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
6C30
 
 | | Mycobacterium smegmatis RimJ (apo form) | | Descriptor: | CHLORIDE ION, GLYCEROL, GNAT family acetyltransferase | | Authors: | Favrot, L, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | Structural Characterization of Mycobacterium smegmatis RimJ, an N-acetyltransferase protein
To Be Published
|
|
6C32
 
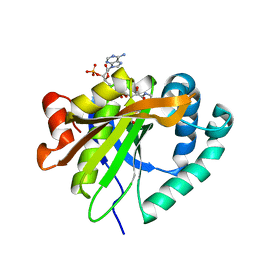 | | Mycobacterium smegmatis RimJ with AcCoA | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, GNAT family protein | | Authors: | Favrot, L, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structural Characterization of Mycobacterium smegmatis RimJ, an N-acetyltransferase protein
To Be Published
|
|
6C5D
 
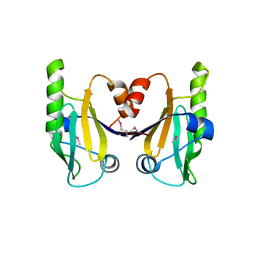 | |
6C37
 
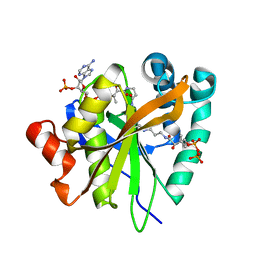 | | Mycobacterium smegmatis RimJ in complex with CoA-disulfide | | Descriptor: | Acetyltransferase, GNAT family protein, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Favrot, L, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Characterization of Mycobacterium smegmatis RimJ, an N-acetyltransferase protein
To Be Published
|
|
3TUD
 
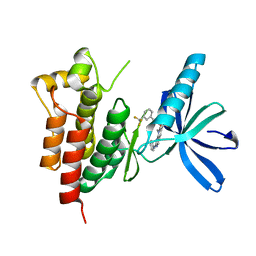 | | Crystal structure of SYK kinase domain with N-(4-methyl-3-(8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl)phenyl)-3-(trifluoromethyl)benzamide | | Descriptor: | N-{4-methyl-3-[8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl]phenyl}-3-(trifluoromethyl)benzamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
6DJQ
 
 | | Vps1 GTPase-BSE fusion complexed with GDP.AlF4- | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Varlakhanova, N.V, Brady, T.M, Tornabene, B.A, Hosford, C.J, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2018-05-25 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
2SOD
 
 | | DETERMINATION AND ANALYSIS OF THE 2 ANGSTROM STRUCTURE OF COPPER, ZINC SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Tainer, J.A, Getzoff, E.D, Richardson, J.S, Richardson, D.C. | | Deposit date: | 1980-03-25 | | Release date: | 1980-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase.
J.Mol.Biol., 160, 1982
|
|
3O45
 
 | | Crystal Structure of 101F Fab Bound to 17-mer Peptide Epitope | | Descriptor: | Fusion glycoprotein F1, Mouse monoclonal antibody 101F 101F Fab heavy chain, Mouse monoclonal antibody 101F 101F Fab light chain, ... | | Authors: | McLellan, J.S, Chen, M, Chang, J.S, Yang, Y, Kim, A, Graham, B.S, Kwong, P.D. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Structure of a Major Antigenic Site on the Respiratory Syncytial Virus Fusion Glycoprotein in Complex with Neutralizing Antibody 101F.
J.Virol., 84, 2010
|
|
3O41
 
 | | Crystal Structure of 101F Fab Bound to 15-mer Peptide Epitope | | Descriptor: | Fusion glycoprotein F1, Mouse monoclonal antibody 101F Fab heavy chain, Mouse monoclonal antibody 101F Fab light chain, ... | | Authors: | McLellan, J.S, Chen, M, Chang, J.S, Yang, Y, Kim, A, Graham, B.S, Kwong, P.D. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Major Antigenic Site on the Respiratory Syncytial Virus Fusion Glycoprotein in Complex with Neutralizing Antibody 101F.
J.Virol., 84, 2010
|
|
3IQA
 
 | | Crystal Structure of BlaC covalently bound with Doripenem | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2009-08-19 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem.
Biochemistry, 49, 2010
|
|
3HT5
 
 | |
1VJK
 
 | | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 | | Descriptor: | molybdopterin converting factor, subunit 1 | | Authors: | Chen, L, Liu, Z.J, Tempel, W, Shah, A, Lee, D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 '
To be published
|
|
