1C5L
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | CALCIUM ION, Hirudin, SODIUM ION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5N
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, Hirudin, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5O
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | BENZAMIDINE, Hirudin, SODIUM ION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5W
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CITRATE ANION, PROTEIN (UROKINASE-TYPE PLASMINOGEN ACTIVATOR) | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5X
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CITRATE ANION, PROTEIN (UROKINASE-TYPE PLASMINOGEN ACTIVATOR) | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5S
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | BENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, PROTEIN (TRYPSIN), ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5Y
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | CITRATE ANION, PROTEIN (UROKINASE-TYPE PLASMINOGEN ACTIVATOR), THIENO[2,3-B]PYRIDINE-2-CARBOXAMIDINE | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
3WIQ
 
 | | Crystal structure of kojibiose phosphorylase complexed with kojibiose | | Descriptor: | Kojibiose phosphorylase, SULFATE ION, alpha-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
3WIR
 
 | | Crystal structure of kojibiose phosphorylase complexed with glucose | | Descriptor: | GLYCEROL, Kojibiose phosphorylase, PHOSPHATE ION, ... | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
1JLZ
 
 | | Solution Structure of a K+-Channel Blocker from the Scorpion Toxin of Tityus cambridgei | | Descriptor: | Tityustoxin alpha-KTx | | Authors: | Wang, I, Wu, S.-H, Chang, H.-K, Shieh, R.-C, Yu, H.-M, Chen, C. | | Deposit date: | 2001-07-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a K(+)-channel blocker from the scorpion Tityus cambridgei.
Protein Sci., 11, 2002
|
|
4O9R
 
 | | Human Smoothened Receptor structure in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Weierstall, U, James, D, White, T.A, Wang, D, Liu, W, Spence, J.C.H, Doak, R.B, Nelson, G, Fromme, P, Fromme, R, Grotjohann, I, Kupitz, C, Zatsepin, N.A, Liu, H, Basu, S, Wacker, D, Han, G.W, Katritch, V, Boutet, S, Messerschmidt, M, Willams, G.J, Koglin, J.E, Seibert, M.M, Klinker, M, Gati, C, Shoeman, R.L, Barty, A, Chapman, H.N, Kirian, R.A, Beyerlein, K.R, Stevens, R.C, Li, D, Shah, S.T.A, Howe, N, Caffrey, M, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography.
Nat Commun, 5, 2014
|
|
1C5Z
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | BENZAMIDINE, CITRATE ANION, PROTEIN (UROKINASE-TYPE PLASMINOGEN ACTIVATOR) | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
4NEK
 
 | | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Geffken, K, Chapman, H.C, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1
To be Published
|
|
1XA1
 
 | | Crystal structure of the sensor domain of BlaR1 from Staphylococcus aureus in its apo form | | Descriptor: | PHOSPHATE ION, PYROPHOSPHATE 2-, Regulatory protein blaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
3ZJ7
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
8VOX
 
 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with MgADP | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VP1
 
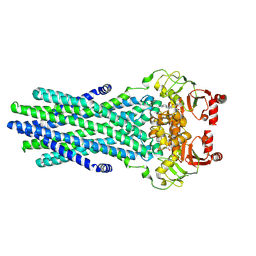 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with ATP | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VOW
 
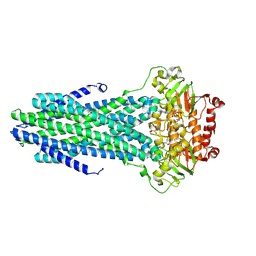 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with MgATP and Vi | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VP8
 
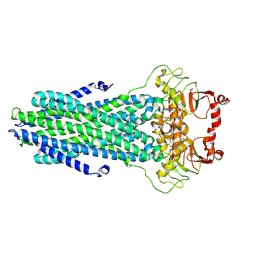 | | Cryo-EM structure of the ABC transporter PCAT1 bound with ATP and Substrate | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VP6
 
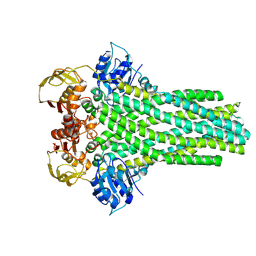 | | Cryo-EM structure of the ABC transporter PCAT1 bound with Mg_class_2 | | Descriptor: | ABC-type bacteriocin transporter | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VPA
 
 | | Cryo-EM structure of the ABC transporter PCAT1 bound with ADP | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VPB
 
 | | Cryo-EM structure of the ABC transporter PCAT1 bound with MgADP_Class_2 | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
9AZL
 
 | | Cryo-EM structure of the ABC transporter PCAT1 bound with MgADP class1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
9BAA
 
 | | Cryo-EM structure of the ABC transporter PCAT1 bound with Mg_class_1 | | Descriptor: | ABC-type bacteriocin transporter | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-04-03 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
4OLQ
 
 | | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium | | Descriptor: | D-MALATE, DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase/isomerase family protein, ... | | Authors: | Szlachta, K, Cooper, D.R, Chapman, H.C, Cymborowski, M.T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-24 | | Release date: | 2014-03-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium
To be Published
|
|
