3ZLG
 
 | | Structure of group A Streptococcal enolase K362A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
3ZLH
 
 | | Structure of group A Streptococcal enolase | | Descriptor: | ENOLASE | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
3ZLF
 
 | | Structure of group A Streptococcal enolase K312A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
4ZM1
 
 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - wild type | | Descriptor: | CITRIC ACID, Chain length determinant protein, MAGNESIUM ION | | Authors: | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | Deposit date: | 2015-05-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
4ZM5
 
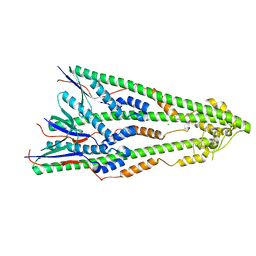 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - A107P mutant | | Descriptor: | CHLORIDE ION, Chain length determinant protein, MAGNESIUM ION | | Authors: | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | Deposit date: | 2015-05-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
4BOF
 
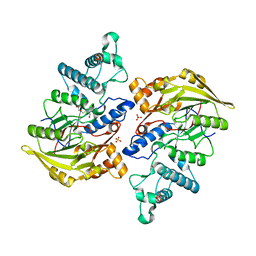 | | Crystal structure of arginine deiminase from group A streptococcus | | Descriptor: | ARGININE DEIMINASE, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Henningham, A, Ericsson, D.J, Langer, K, Casey, L, Jovcevski, B, Chhatwal, G.S, Aquilina, J.A, Batzloff, M.R, Kobe, B, Walker, M.J. | | Deposit date: | 2013-05-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-informed design of an enzymatically inactive vaccine component for group A Streptococcus.
MBio, 4, 2013
|
|
4C7M
 
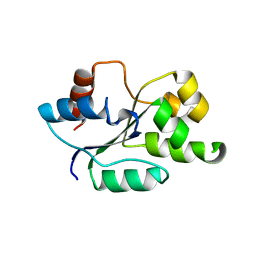 | | The crystal structure of TcpB or BtpA TIR domain | | Descriptor: | Toll/interleukin-1 receptor domain-containing protein | | Authors: | Alaidarous, M, Ve, T, Casey, L.W, Valkov, E, Ullah, M.O, Schembri, M.A, Mansell, A, Sweet, M.J, Kobe, B. | | Deposit date: | 2013-09-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Mechanism of bacterial interference with TLR4 signaling by Brucella Toll/interleukin-1 receptor domain-containing protein TcpB.
J.Biol.Chem., 289, 2014
|
|
6O0V
 
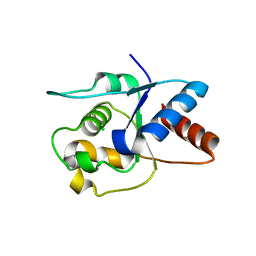 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0T
 
 | | Crystal structure of selenomethionine labelled tandem SAM domains (L446M:L505M:L523M mutant) from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0W
 
 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0S
 
 | | Crystal structure of the tandem SAM domains from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O1B
 
 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0R
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with glycerol | | Descriptor: | GLYCEROL, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0U
 
 | | Crystal structure of the TIR domain H685A mutant from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0Q
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with ribose | | Descriptor: | CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1, beta-D-ribofuranose | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
