2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
6FF9
 
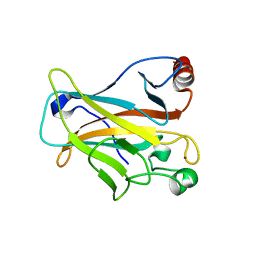 | | Mutant R280K of human P53 | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Trovao, F.G, Gomes, A.S, Pinheiro, B, Carvalho, A.L, Romao, M.J. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the R280K Mutant of Human p53 Explains the Loss of DNA Binding.
Int J Mol Sci, 19, 2018
|
|
6G0V
 
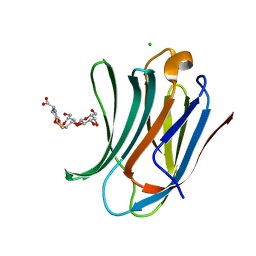 | | Human Galectin-3 in complex with a TF tumor-associated antigen mimetic | | Descriptor: | (3~{R},5~{R},6~{S},7~{S},8~{R},13~{S})-5-(hydroxymethyl)-7-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-oxidanyl-11-oxidanylidene-2,4-dioxa-9-thia-12-azatricyclo[8.4.0.0^{3,8}]tetradec-1(10)-ene-13-carboxylic acid, CHLORIDE ION, Galectin-3 | | Authors: | Trovao, F.G, Santarsia, S, Carvalho, A.L. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular Recognition of a Thomsen-Friedenreich Antigen Mimetic Targeting Human Galectin-3.
ChemMedChem, 13, 2018
|
|
6FOP
 
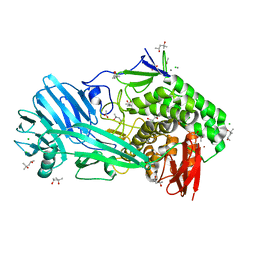 | | Glycoside hydrolase family 81 from Clostridium thermocellum (CtLam81A), Mutant E515A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Correia, M.A.S.C, Carvalho, A.L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel insights into the degradation of beta-1,3-glucans by the cellulosome of Clostridium thermocellum revealed by structure and function studies of a family 81 glycoside hydrolase.
Int.J.Biol.Macromol., 117, 2018
|
|
3UL4
 
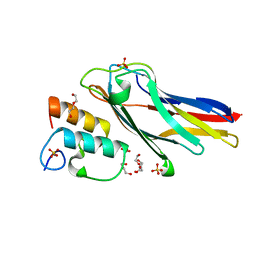 | | Crystal structure of Coh-OlpA(Cthe_3080)-Doc918(Cthe_0918) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome enzyme, dockerin type I, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
5K39
 
 | | THE TYPE II COHESIN DOCKERIN COMPLEX FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin module from a protein of unknown function | | Authors: | Viegas, A, Pinheiro, B, Bras, J.L.A, Romao, M.J, Alves, V, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
5N5P
 
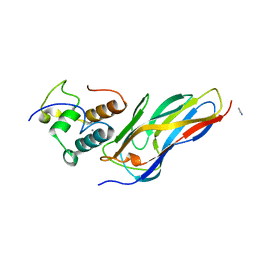 | | Crystal structure of Ruminococcus flavefaciens' type III complex containing the fifth cohesin from scaffoldin B and the dockerin from scaffoldin A | | Descriptor: | ACETONITRILE, CALCIUM ION, Putative cellulosomal scaffoldin protein | | Authors: | Bule, P, Carvalho, A.L, Najmudin, S, Fontes, C.M.G.A. | | Deposit date: | 2017-02-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Higher order scaffoldin assembly in Ruminococcus flavefaciens cellulosome is coordinated by a discrete cohesin-dockerin interaction.
Sci Rep, 8, 2018
|
|
5M2O
 
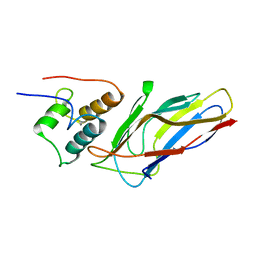 | | R. flavefaciens' third ScaB cohesin in complex with a group 1 dockerin | | Descriptor: | CALCIUM ION, Group I Dockerin, Putative cellulosomal scaffoldin protein | | Authors: | Bule, P, Najmudin, S, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes.
Sci Rep, 7, 2017
|
|
5LU3
 
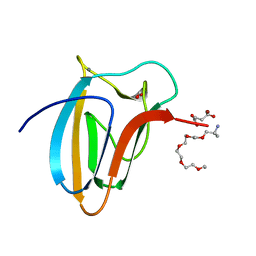 | | The Structure of Spirochaeta thermophila CBM64 | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Correia, M.A.S, Romao, M.J, Carvalho, A.L. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stability and Ligand Promiscuity of Type A Carbohydrate-binding Modules Are Illustrated by the Structure of Spirochaeta thermophila StCBM64C.
J. Biol. Chem., 292, 2017
|
|
5M2S
 
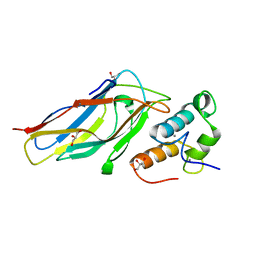 | | R. flavefaciens' third ScaB cohesin in complex with a group 1 dockerin | | Descriptor: | CALCIUM ION, Doc8: Type I dockerin repeat domain from family 9 glycoside hydrolase WP_009982745[Ruminococcus flavefaciens], GLYCEROL, ... | | Authors: | Bule, P, Najmudin, S, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes.
Sci Rep, 7, 2017
|
|
5M0Y
 
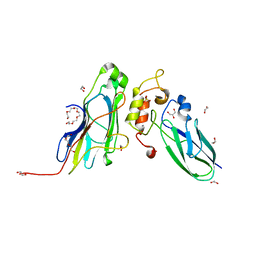 | | Crystal Structure of the CohScaA-XDocCipB type II complex from Clostridium thermocellum at 1.5Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cellulosome anchoring protein cohesin region, ... | | Authors: | Pinheiro, B.A, Bras, J.L, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-06 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
7R1L
 
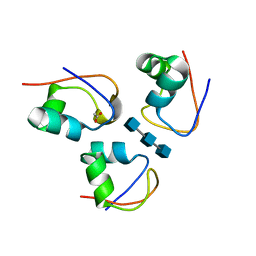 | | Clostridium thermocellum CtCBM50 structure in complex with beta-1,4-GlcNAc trisaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Spore coat assembly protein SafA | | Authors: | Ribeiro, D.O, Costa, R, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unravelling Clostridium thermocellum LysM domains: a molecular view on the cooperative recognition of chitin and peptidoglycan
To Be Published
|
|
4AYZ
 
 | | X-ray Structure of human SOUL | | Descriptor: | HEME-BINDING PROTEIN 2 | | Authors: | Freire, F, Carvalho, A.L, Aveiro, S.S, Charbonnier, P, Moulis, J.M, Romao, M.J, Goodfellow, B.J, Macedo, A.L. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Human Soul: A Heme-Binding or a Bh3 Domain-Containing Protein
To be Published
|
|
4B0Y
 
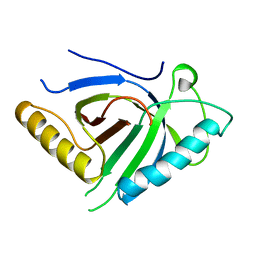 | | Determination of X-ray Structure of human SOUL by Molecular Replacement | | Descriptor: | HEME-BINDING PROTEIN 2 | | Authors: | Freire, F, Carvalho, A.L, Aveiro, S.S, Charbonnier, P, Moulis, J.M, Romao, M.J, Goodfellow, B.J, Macedo, A.L. | | Deposit date: | 2012-07-06 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Human Soul: A Heme-Binding or a Bh3 Domain-Containing Protein
To be Published
|
|
1CFW
 
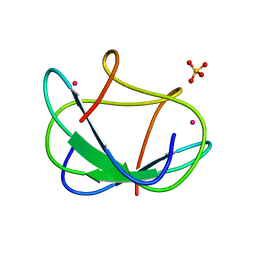 | | GA-SUBSTITUTED DESULFOREDOXIN | | Descriptor: | GALLIUM (III) ION, PROTEIN (DESULFOREDOXIN), SULFATE ION | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Moura, I, Moura, J.J.G, Rusnak, F, Romao, M.J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
1DCD
 
 | | DESULFOREDOXIN COMPLEXED WITH CD2+ | | Descriptor: | CADMIUM ION, PROTEIN (DESULFOREDOXIN) | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Romao, M.J. | | Deposit date: | 1999-03-20 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
1DHG
 
 | | HG-SUBSTITUTED DESULFOREDOXIN | | Descriptor: | MERCURY (II) ION, PROTEIN (DESULFOREDOXIN) | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Moura, I, Moura, J.J.G, Rusnak, F, Romao, M.J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
1SFP
 
 | | CRYSTAL STRUCTURE OF ACIDIC SEMINAL FLUID PROTEIN (ASFP) AT 1.9 A RESOLUTION: A BOVINE POLYPEPTIDE FROM THE SPERMADHESIN FAMILY | | Descriptor: | ASFP | | Authors: | Romao, M.J, Kolln, I, Dias, J.M, Carvalho, A.L, Romero, A, Varela, P.F, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
1SPP
 
 | | THE CRYSTAL STRUCTURES OF TWO MEMBERS OF THE SPERMADHESIN FAMILY REVEAL THE FOLDING OF THE CUB DOMAIN | | Descriptor: | MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-I, MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-II | | Authors: | Romero, A, Romao, M.J, Varela, P.F, Kolln, I, Dias, J.M, Carvalho, A.L, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-19 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
4AO1
 
 | | High resolution crystal structure of bovine pancreatic ribonuclease crystallized using ionic liquid | | Descriptor: | CHLORIDE ION, RIBONUCLEASE PANCREATIC, SULFATE ION | | Authors: | Mukhopadhyay, A, Carvalho, A.L, Kowacz, M, Romao, M.J. | | Deposit date: | 2012-03-23 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Hofmeister Effects of Ionic Liquids in Protein Crystallization: Direct and Water-Mediated Interactions
Cryst.Eng.Comm., 14, 2012
|
|
4AGA
 
 | | Hofmeister effects of ionic liquids in protein crystallization: direct and water-mediated interactions | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Mukhopadhyay, A, Carvalho, A.L, Romao, M.J. | | Deposit date: | 2012-01-25 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hofmeister Effects of Ionic Liquids in Protein Crystallization: Direct and Water-Mediated Interactions
Cryst.Eng.Comm., 14, 2012
|
|
4DH2
 
 | | Crystal structure of Coh-OlpC(Cthe_0452)-Doc435(Cthe_0435) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin type 1, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
7BLH
 
 | |
8AJY
 
 | | Ruminococcus flavefaciens Cohesin-Dockerin structure: dockerin from ScaH adaptor scaffoldin in complex with the cohesin from ScaE anchoring scaffoldin | | Descriptor: | CALCIUM ION, Cell-wall anchoring protein, Dockerin from ScaH, ... | | Authors: | Alves, V.D, Bule, P, Fontes, C.M.G.A, Carvalho, A.L.M, Najmudin, S, Duarte, M. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-function studies can improve binding affinity of cohesin-dockerin interactions for multi-protein assemblies.
Int.J.Biol.Macromol., 224, 2023
|
|
