6TWG
 
 | | Solution structure of antimicrobial peptide, crabrolin Plus in the presence of Lipopolysaccharide | | Descriptor: | Crabrolin Plus, mutant of Crabrolin peptide | | Authors: | Cantini, F, Bouchemal, N, Savarin, P, Sette, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Effect of positive charges in the structural interaction of crabrolin isoforms with lipopolysaccharide.
J.Pept.Sci., 26, 2020
|
|
6XYV
 
 | |
7A58
 
 | |
1YS5
 
 | | Solution structure of the antigenic domain of GNA1870 of Neisseria meningitidis | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Savino, S, Masignani, V, Pizza, M, Scarselli, M, Swennen, E, Romagnoli, G, Veggi, D, Banci, L, Rappuoli, R. | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis
J.Biol.Chem., 281, 2006
|
|
2K4B
 
 | | CopR Repressor Structure | | Descriptor: | Transcriptional regulator | | Authors: | Cantini, F, Banci, L, Magnani, D, Solioz, M. | | Deposit date: | 2008-06-03 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The copper-responsive repressor CopR of Lactococcus lactis is a 'winged helix' protein.
Biochem.J., 417, 2009
|
|
2KC0
 
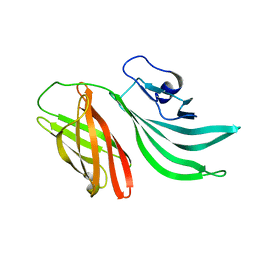 | | Solution structure of the factor H binding protein | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Veggi, D, Dragonetti, S, Savino, S, Scarselli, M, Romagnoli, G, Pizza, M, Banci, L, Rappuoli, R. | | Deposit date: | 2008-12-13 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Factor H-binding Protein, a Survival Factor and Protective Antigen of Neisseria meningitidis
J.Biol.Chem., 284, 2009
|
|
6FFK
 
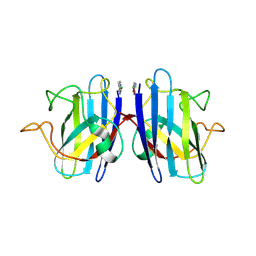 | | Human apo-SOD1 bound to PtCl2(1R,2R-1,4-DACH | | Descriptor: | PtCl2(1(R),2(R)-DACH), Superoxide dismutase [Cu-Zn] | | Authors: | Calderone, V, Nativi, C, Cantini, F, Di Cesare Mannelli, L. | | Deposit date: | 2018-01-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Interaction of Half Oxa-/Halfcis-Platin Complex with Human Superoxide Dismutase and Induced Reduction of Neurotoxicity.
ACS Med Chem Lett, 9, 2018
|
|
7A4L
 
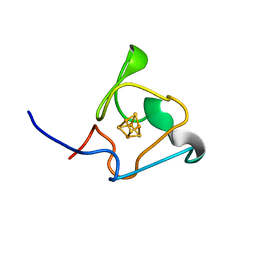 | | PRE-only solution structure of the Iron-Sulfur protein PioC from Rhodopseudomonas palustris TIE-1 | | Descriptor: | IRON/SULFUR CLUSTER, PioC | | Authors: | Trindade, I, Invernici, M, Cantini, F, Louro, R, Piccioli, M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PRE-driven protein NMR structures: an alternative approach in highly paramagnetic systems.
Febs J., 288, 2021
|
|
2KIJ
 
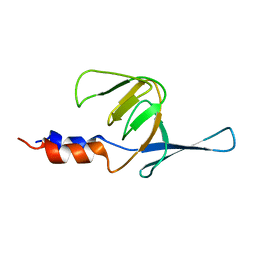 | | Solution structure of the Actuator domain of the copper-transporting ATPase ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Nushi, F, Natile, G, Rosato, A. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the actuator domain of ATP7A and ATP7B, the Menkes and Wilson disease proteins
Biochemistry, 48, 2009
|
|
2L4O
 
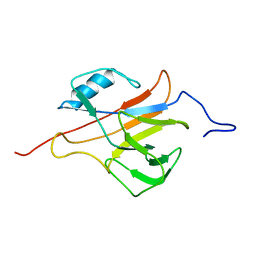 | | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain | | Descriptor: | Cell wall surface anchor family protein | | Authors: | Gentile, M.A, Melchiorre, S, Emolo, C, Moschioni, M, Gianfaldoni, C, Pancotto, L, Ferlenghi, I, Scarselli, M, Pansegrau, W, Veggi, D, Merola, M, Cantini, F, Ruggiero, P, Banci, L, Masignani, V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain: in vivo protection and implications for the molecular mechanisms of pilus polymerization
To be Published
|
|
2KMV
 
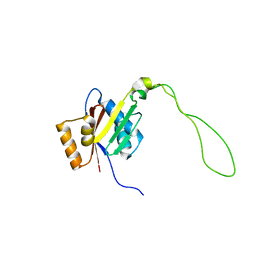 | | Solution structure of the nucleotide binding domain of the human Menkes protein in the ATP-free form | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Inagaki, S, Migliardi, M, Rosato, A. | | Deposit date: | 2009-08-05 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A.
J.Biol.Chem., 2009
|
|
8B0T
 
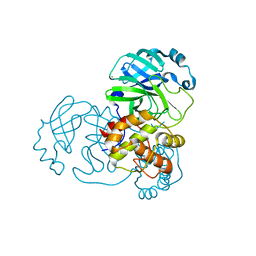 | | SARS-CoV-2 Main Protease adduct with Au(PEt3)Br | | Descriptor: | 3C-like proteinase nsp5, GOLD ION | | Authors: | Massai, L, Grifagni, D, De Santis, A, Geri, A, Calderone, V, Cantini, F, Banci, L, Messori, L. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gold-Based Metal Drugs as Inhibitors of Coronavirus Proteins: The Inhibition of SARS-CoV-2 Main Protease by Auranofin and Its Analogs.
Biomolecules, 12, 2022
|
|
8B0S
 
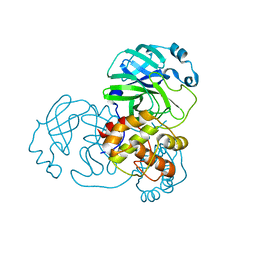 | | SARS-COV-2 Main Protease adduct with Au(NHC)Cl | | Descriptor: | 3C-like proteinase nsp5, GOLD ION | | Authors: | Massai, L, Grifagni, D, Desantis, A, Geri, A, Calderone, V, Cantini, F, Messori, L, Banci, L. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Gold-Based Metal Drugs as Inhibitors of Coronavirus Proteins: The Inhibition of SARS-CoV-2 Main Protease by Auranofin and Its Analogs.
Biomolecules, 12, 2022
|
|
8S6F
 
 | | A Structural Investigation of the Interaction between a GC-376-Based Peptidomimetic PROTAC and Its Precursor with the Viral Main Protease of Coxsackievirus B3 | | Descriptor: | Protease 3C, methyl (4~{S})-4-[[(2~{S})-4-methyl-2-(phenylmethoxycarbonylamino)pentanoyl]amino]-5-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | De Santis, A, Grifagni, D, Orsetti, A, Lenci, E, Rosato, A, D'Onofrio, M, Trabocchi, A, Ciofi-Baffoni, S, Cantini, F, Calderone, V. | | Deposit date: | 2024-02-27 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Structural Investigation of the Interaction between a GC-376-Based Peptidomimetic PROTAC and Its Precursor with the Viral Main Protease of Coxsackievirus B3.
Biomolecules, 14, 2024
|
|
1ON4
 
 | | Solution structure of soluble domain of Sco1 from Bacillus Subtilis | | Descriptor: | Sco1 | | Authors: | Balatri, E, Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-02-27 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Sco1: A Thioredoxin-like Protein Involved in Cytochrome c Oxidase Assembly
STRUCTURE, 11, 2003
|
|
1KMG
 
 | | The Solution Structure Of Monomeric Copper-free Superoxide Dismutase | | Descriptor: | Superoxide Dismutase, ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Onofrio, M, Viezzoli, M.S. | | Deposit date: | 2001-12-15 | | Release date: | 2002-10-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of copper-free SOD: The protein before binding copper.
Protein Sci., 11, 2002
|
|
2MPF
 
 | | Solution structure human HCN2 CNBD in the cAMP-unbound state | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Saponaro, A, Pauleta, S.R, Cantini, F, Matzapetakis, M, Hammann, C, Banci, L, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7NXH
 
 | | Structure of SARS-CoV2 NSP5 (3C-like proteinase) determined in-house | | Descriptor: | 3C-like proteinase | | Authors: | Calderone, V, Grifagni, D, Cantini, F, Fragai, M, Banci, L. | | Deposit date: | 2021-03-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 M pro inhibition by a zinc ion: structural features and hints for drug design.
Chem.Commun.(Camb.), 57, 2021
|
|
7NWX
 
 | | SARS-COV2 NSP5 in the presence of Zn2+ | | Descriptor: | Replicase polyprotein 1a, ZINC ION | | Authors: | Calderone, V, Grifagni, D, Cantini, F, Fragai, M, Banci, L. | | Deposit date: | 2021-03-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 M pro inhibition by a zinc ion: structural features and hints for drug design.
Chem.Commun.(Camb.), 57, 2021
|
|
2N6J
 
 | | Solution structure of Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile | | Descriptor: | ZINC ION, Zinc metalloprotease Zmp1 | | Authors: | Banci, L, Cantini, F, Scarselli, M, Rubino, J.T, Martinelli, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of zinc-bound Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile.
J.Biol.Inorg.Chem., 21, 2016
|
|
2RSQ
 
 | | Copper(I) loaded form of the first domain of the human copper chaperone for SOD1, CCS | | Descriptor: | COPPER (I) ION, Copper chaperone for superoxide dismutase | | Authors: | Banci, L, Bertini, I, Cantini, F, Kozyreva, T, Rubino, J.T. | | Deposit date: | 2012-05-15 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Copper(I) loaded form of the first domain of the human copper chaperone for SOD1, CCS
To be Published
|
|
2ROP
 
 | | Solution structure of domains 3 and 4 of human ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Banci, L, Bertini, I, Cantini, F, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2008-04-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1
Biochemistry, 47, 2008
|
|
1SP0
 
 | | Solution Structure of apoCox11 | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1TL5
 
 | | Solution structure of apoHAH1 | | Descriptor: | Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
1TL4
 
 | | Solution structure of Cu(I) HAH1 | | Descriptor: | COPPER (I) ION, Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
