1L5I
 
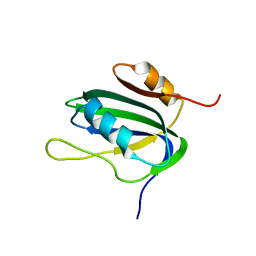 | | 30-CONFORMER NMR ENSEMBLE OF THE N-TERMINAL, DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN FROM A GEMINIVIRUS (TOMATO YELLOW LEAF CURL VIRUS-SARDINIA) | | Descriptor: | Rep protein | | Authors: | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | Deposit date: | 2002-03-07 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L2M
 
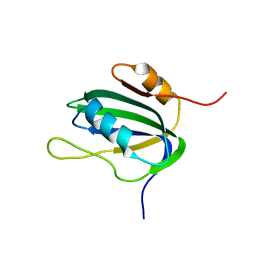 | | Minimized Average Structure of the N-terminal, DNA-binding domain of the replication initiation protein from a geminivirus (Tomato yellow leaf curl virus-Sardinia) | | Descriptor: | Rep protein | | Authors: | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | Deposit date: | 2002-02-22 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GH5
 
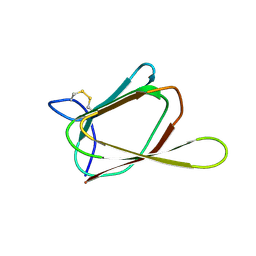 | | ANTIFUNGAL PROTEIN FROM STREPTOMYCES TENDAE TU901, NMR AVERAGE STRUCTURE | | Descriptor: | ANTIFUNGAL PROTEIN | | Authors: | Campos-Olivas, R, Bormann, C, Hoerr, I, Jung, G, Gronenborn, A.M. | | Deposit date: | 2000-11-04 | | Release date: | 2001-03-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics and chitin binding of the anti-fungal protein from Streptomyces tendae TU901.
J.Mol.Biol., 308, 2001
|
|
1G6E
 
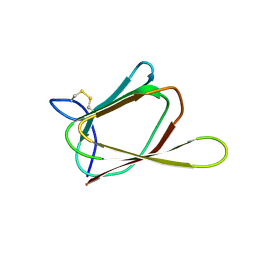 | | ANTIFUNGAL PROTEIN FROM STREPTOMYCES TENDAE TU901, 30-CONFORMERS ENSEMBLE | | Descriptor: | ANTIFUNGAL PROTEIN | | Authors: | Campos-Olivas, R, Bormann, C, Hoerr, I, Jung, G, Gronenborn, A.M. | | Deposit date: | 2000-11-04 | | Release date: | 2001-03-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics and chitin binding of the anti-fungal protein from Streptomyces tendae TU901.
J.Mol.Biol., 308, 2001
|
|
1D1D
 
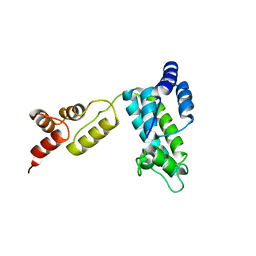 | |
1AFP
 
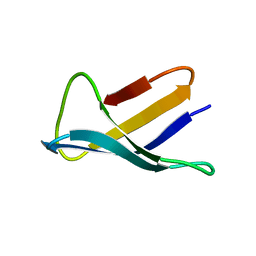 | | SOLUTION STRUCTURE OF THE ANTIFUNGAL PROTEIN FROM ASPERGILLUS GIGANTEUS. EVIDENCE FOR DISULPHIDE CONFIGURATIONAL ISOMERISM | | Descriptor: | ANTIFUNGAL PROTEIN FROM ASPERGILLUS GIGANTEUS | | Authors: | Campos-Olivas, R, Bruix, M, Santoro, J, Lacadena, J, Del Pozo, A.M, Gavilanes, J.G, Rico, M. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the antifungal protein from Aspergillus giganteus: evidence for cysteine pairing isomerism.
Biochemistry, 34, 1995
|
|
2N89
 
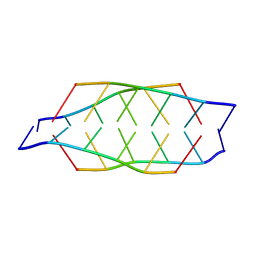 | | Tetrameric i-motif structure of dT-dC-dC-CFL-CFL-dC at acidic pH | | Descriptor: | DNA (5'-D(*TP*CP*CP*(CFL)P*(CFL)P*C)-3') | | Authors: | Abou-Assi, H, Harkness, R.W, Martin-Pintado, N, Wilds, C.J, Campos-Olivas, R, Mittermaier, A.K, Gonzalez, C, Damha, M.J. | | Deposit date: | 2015-10-09 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stabilization of i-motif structures by 2'-beta-fluorination of DNA.
Nucleic Acids Res., 44, 2016
|
|
2MQK
 
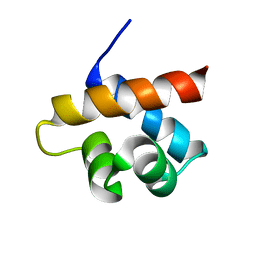 | |
1DE3
 
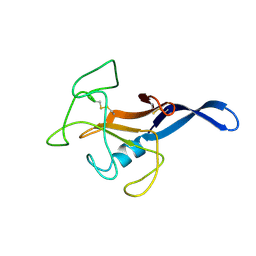 | | SOLUTION STRUCTURE OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | RIBONUCLEASE ALPHA-SARCIN | | Authors: | Perez-Canadillas, J.M, Campos-Olivas, R, Santoro, J, Lacadena, J, Martinez del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 1999-11-12 | | Release date: | 2000-06-21 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity.
J.Mol.Biol., 299, 2000
|
|
4CJA
 
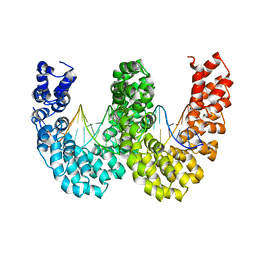 | | BurrH DNA-binding protein from Burkholderia rhizoxinica in complex with its target DNA | | Descriptor: | 5'-D(*DTP*AP*TP*AP*AP*CP*GP*TP*AP*TP*TP*TP*GP*CP *TP*TP*CP*TP*CP*TP*TP*AP*AP)-3', 5'-D(*DTP*TP*AP*AP*GP*AP*GP*AP*AP*GP*CP*AP*AP*DP *TP*AP*CP*GP*TP*TP*AP*TP*AP)-3', BURRH | | Authors: | Stella, S, Molina, R, Lopez-Mendez, B, Campos-Olivas, R, Duchateau, P, Montoya, G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Bud, a Helix-Loop-Helix DNA-Binding Domain for Genome Modification
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CJ9
 
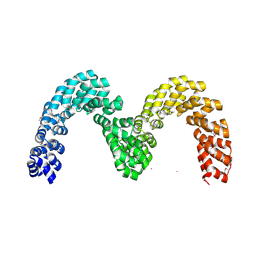 | | BurrH DNA-binding protein from Burkholderia rhizoxinica in its apo form | | Descriptor: | BURRH, SELENIUM ATOM | | Authors: | Stella, S, Molina, R, Lopez-Mendez, B, Campos-Olivas, R, Duchateau, P, Montoya, G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Bud, a Helix-Loop-Helix DNA-Binding Domain for Genome Modification
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1PU1
 
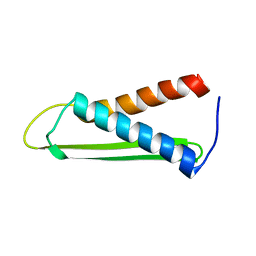 | | Solution structure of the Hypothetical protein mth677 from Methanothermobacter Thermautotrophicus | | Descriptor: | Hypothetical protein MTH677 | | Authors: | Blanco, F.J, Yee, A, Campos-Olivas, R, Devos, D, Valencia, A, Arrowsmith, C.H, Rico, M. | | Deposit date: | 2003-06-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical protein Mth677 from Methanobacterium thermoautotrophicum: A novel {alpha}+{beta} fold
Protein Sci., 13, 2004
|
|
1QRJ
 
 | | Solution structure of htlv-i capsid protein | | Descriptor: | HTLV-I CAPSID PROTEIN | | Authors: | Khorasanizadeh, S, Campos-Olivas, R, Clark, C.A, Summers, M.F. | | Deposit date: | 1999-06-14 | | Release date: | 1999-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific 1H, 13C and 15N chemical shift assignment and secondary structure of the HTLV-I capsid protein.
J.Biomol.NMR, 14, 1999
|
|
2KG4
 
 | | Three-dimensional structure of human Gadd45alpha in solution by NMR | | Descriptor: | Growth arrest and DNA-damage-inducible protein GADD45 alpha | | Authors: | Sanchez, R, Pantoja-Uceda, D, Prieto, J, Diercks, T, Campos-Olivas, R, Blanco, F.J. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase
J.Biol.Chem., 285, 2010
|
|
2HWT
 
 | |
2HW0
 
 | | NMR Solution Structure of the nuclease domain from the Replicator Initiator Protein from porcine circovirus PCV2 | | Descriptor: | Replicase | | Authors: | Vega-Rocha, S, Byeon, I.L, Gronenborn, B, Gronenborn, A.M, Campos-Olivas, R. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2
J.Mol.Biol., 367, 2007
|
|
5FT8
 
 | | Crystal structure of the complex between the cysteine desulfurase CsdA and the sulfur-acceptor CsdE in the persulfurated state at 2.50 Angstroem resolution | | Descriptor: | Cysteine desulfurase CsdA, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
5FT5
 
 | | Crystal structure of the cysteine desulfurase CsdA (persulfurated) from Escherichia coli at 2.384 Angstroem resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Quintana, J.F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
5FT6
 
 | | Crystal structure of the cysteine desulfurase CsdA (S-sulfonic acid) from Escherichia coli at 2.050 Angstroem resolution | | Descriptor: | CYSTEINE DESULFURASE CSDA, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
5FT4
 
 | | Crystal structure of the cysteine desulfurase CsdA from Escherichia coli at 1.996 Angstroem resolution | | Descriptor: | CITRIC ACID, CYSTEINE DESULFURASE CSDA, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Quintana, J.F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-12-21 | | Last modified: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | The Mechanism of Sulfur Transfer Across Protein- Protein Interfaces: The Csd Model
Acs Catalysis, 6, 2016
|
|
