1LOD
 
 | |
1KXT
 
 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
1LOF
 
 | |
1LK7
 
 | | Structure of D-Ribose-5-Phosphate Isomerase from in complex with phospho-erythronic acid | | Descriptor: | CHLORIDE ION, D-4-PHOSPHOERYTHRONIC ACID, D-Ribose-5-Phosphate Isomerase, ... | | Authors: | Ishikawa, K, Matsui, I, Payan, F, Cambillau, C, Ishida, H, Kawarabayasi, Y, Kikuchi, H, Roussel, A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Hyperthermostable D-Ribose-5-Phosphate Isomerase from Pyrococcus horikoshii Characterization and Three-Dimensional Structure
STRUCTURE, 10, 2002
|
|
1NZJ
 
 | | Crystal Structure and Activity Studies of Escherichia Coli Yadb ORF | | Descriptor: | Hypothetical protein yadB, ZINC ION | | Authors: | Campanacci, V, Kern, D.Y, Becker, H.D, Spinelli, S, Valencia, C, Vincentelli, R, Pagot, F, Bignon, C, Giege, R, Cambillau, C. | | Deposit date: | 2003-02-18 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity.
J.Mol.Biol., 337, 2004
|
|
3D8L
 
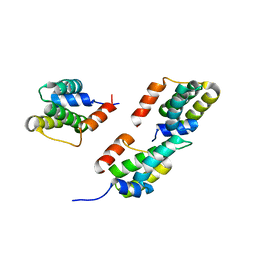 | | Crystal structure of ORF12 from the lactococcus lactis bacteriophage p2 | | Descriptor: | ORF12 | | Authors: | Siponen, M.I, Spinelli, S, Lichiere, J, Moineau, S, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-05-23 | | Release date: | 2009-04-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of ORF12 from Lactococcus lactis phage p2 identifies a tape measure protein chaperone
J.Bacteriol., 191, 2009
|
|
1OKS
 
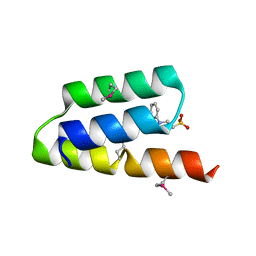 | | Crystal structure of the measles virus phosphoprotein XD domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Johansson, K, Bourhis, J.-M, Campanacci, V, Cambillau, C, Canard, B, Longhi, S. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Measles Virus Phosphoprotein Domain Responsible for the Induced Folding of the C-Terminal Domain of the Nucleoprotein
J.Biol.Chem., 278, 2003
|
|
1K19
 
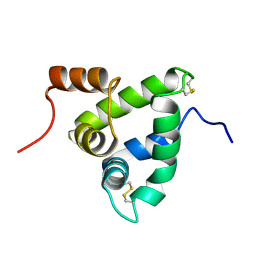 | | NMR Solution Structure of the Chemosensory Protein CSP2 from Moth Mamestra brassicae | | Descriptor: | Chemosensory Protein CSP2 | | Authors: | Mosbah, A, Campanacci, V, Lartigue, A, Tegoni, M, Cambillau, C, Darbon, H. | | Deposit date: | 2001-09-24 | | Release date: | 2002-12-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a chemosensory protein from the moth Mamestra brassicae
BIOCHEM.J., 369, 2003
|
|
3DJW
 
 | | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1 | | Descriptor: | ORF99 | | Authors: | Goulet, A, Spinelli, S, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1
Protein Sci., 18, 2009
|
|
1NIR
 
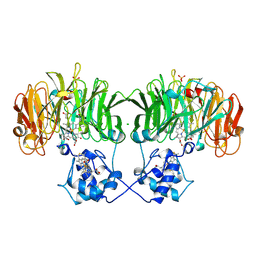 | | OXYDIZED NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | CHLORIDE ION, HEME C, HEME D, ... | | Authors: | Nurizzo, D, Tegoni, M, Cambillau, C. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-terminal arm exchange is observed in the 2.15 A crystal structure of oxidized nitrite reductase from Pseudomonas aeruginosa.
Structure, 5, 1997
|
|
1HLG
 
 | | CRYSTAL STRUCTURE OF HUMAN GASTRIC LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIPASE, ... | | Authors: | Roussel, A, Canaan, S, Verger, R, Cambillau, C. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human gastric lipase and model of lysosomal acid lipase, two lipolytic enzymes of medical interest.
J.Biol.Chem., 274, 1999
|
|
1O8C
 
 | | CRYSTAL STRUCTURE OF E. COLI K-12 YHDH WITH BOUND NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, YHDH | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-26 | | Release date: | 2004-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia Coli Yhdh, a Putative Quinone Oxidoreductase
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1LK5
 
 | | Structure of the D-Ribose-5-Phosphate Isomerase from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, D-Ribose-5-Phosphate Isomerase, SODIUM ION | | Authors: | Ishikawa, K, Matsui, I, Payan, F, Cambillau, C, Ishida, H, Kawarabayasi, Y, Kikuchi, H, Roussel, A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A hyperthermostable D-ribose-5-phosphate isomerase from Pyrococcus horikoshii characterization and three-dimensional structure.
Structure, 10, 2002
|
|
1LOC
 
 | |
1LGC
 
 | | INTERACTION OF A LEGUME LECTIN WITH THE N2 FRAGMENT OF HUMAN LACTOTRANSFERRIN OR WITH THE ISOLATED BIANTENNARY GLYCOPEPTIDE: ROLE OF THE FUCOSE MOIETY | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DIPEPTIDE, ... | | Authors: | Bourne, Y, Cambillau, C. | | Deposit date: | 1994-01-07 | | Release date: | 1994-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a legume lectin complexed with the human lactotransferrin N2 fragment, and with an isolated biantennary glycopeptide: role of the fucose moiety.
Structure, 2, 1994
|
|
1NNO
 
 | | CONFORMATIONAL CHANGES OCCURRING UPON NO BINDING IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Nurizzo, D, Tegoni, M, Cambillau, C. | | Deposit date: | 1998-07-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
1QD0
 
 | | CAMELID HEAVY CHAIN VARIABLE DOMAINS PROVIDE EFFICIENT COMBINING SITES TO HAPTENS | | Descriptor: | 3-HYDROXY-7-(4-{1-[2-HYDROXY-3-(2-HYDROXY-5-SULFO-PHENYLAZO)-BENZYL]-2-SULFO-ETHYLAMINO}-[1,2,5]TRIAZIN-2-YLAMINO)-2-(2-HYDROXY-5-SULFO-PHENYLAZO)-NAPTHALENE-1,8-DISULFONIC ACID, COPPER (II) ION, VHH-R2 ANTI-RR6 ANTIBODY | | Authors: | Spinelli, S, Frenken, L.G.J, Hermans, P, Verrips, T, Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 1999-07-08 | | Release date: | 2000-07-19 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Camelid heavy-chain variable domains provide efficient combining sites to haptens.
Biochemistry, 39, 2000
|
|
1AGY
 
 | |
3D8M
 
 | | Crystal structure of a chimeric receptor binding protein from lactococcal phages subspecies TP901-1 and p2 | | Descriptor: | Baseplate protein, Receptor binding protein | | Authors: | Siponen, M.I, Blangy, S, Spinelli, S, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-05-23 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of a chimeric receptor binding protein constructed from two lactococcal phages
J.Bacteriol., 191, 2009
|
|
1P4B
 
 | | Three-Dimensional Structure Of a Single Chain Fv Fragment Complexed With The peptide GCN4(7P-14P). | | Descriptor: | Antibody Variable heavy chain, Antibody Variable light chain, GCN4(7P-14P) peptide | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-22 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
1P4I
 
 | | Crystal Structure of scFv against peptide GCN4 | | Descriptor: | ANTIBODY VARIABLE LIGHT CHAIN, antibody variable heavy chain | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
1PT7
 
 | | Crystal structure of the apo-form of the yfdW gene product of E. coli | | Descriptor: | GLYCEROL, Hypothetical protein yfdW, PHOSPHATE ION | | Authors: | Gruez, A, Roig-Zamboni, V, Valencia, C, Campanacci, V, Cambillau, C. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the Escherichia coli yfdW gene product reveals a New fold of two interlaced rings identifying a wide family of CoA transferases.
J.Biol.Chem., 278, 2003
|
|
1PT8
 
 | | Crystal structure of the yfdW gene product of E. coli, in complex with oxalate and acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, Hypothetical protein yfdW, ... | | Authors: | Gruez, A, Roig-Zamboni, V, Valencia, C, Campanacci, V, Cambillau, C. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the Escherichia coli yfdW gene product reveals a new fold of two interlaced rings identifying a wide family of CoA transferases.
J.Biol.Chem., 278, 2003
|
|
1PT5
 
 | | Crystal structure of gene yfdW of E. coli | | Descriptor: | ACETYL COENZYME *A, Hypothetical protein yfdW | | Authors: | Gruez, A, Roig-Zamboni, V, Valencia, C, Campanacci, V, Cambillau, C. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Escherichia coli YfdW Gene Product Reveals a New Fold of Two Interlaced Rings Identifying a Wide Family of CoA Transferases
J.Biol.Chem., 278, 2003
|
|
3FAJ
 
 | | Structure of the structural protein P131 of the archaeal virus Acidianus Two-tailed virus (ATV) | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein | | Authors: | Goulet, A, Vestergaard, G, Scheele, U, Campanacci, V, Garrett, R.A, Cambillau, C. | | Deposit date: | 2008-11-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the structural protein P131 of the archaeal virus Acidianus Two-tailed virus (ATV)
To be Published
|
|
