5XK9
 
 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with GSPP and DMAPP | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XK7
 
 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with DMAPP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XK8
 
 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with GPP | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, Undecaprenyl diphosphate synthase | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XWZ
 
 | | Crystal structure of a lactonase from Cladophialophora bantiana | | Descriptor: | GLYCEROL, MALONATE ION, SODIUM ION, ... | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-06-30 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a novel zearalenone hydrolase from Cladophialophora bantiana
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5XSL
 
 | |
8HJT
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
1BC4
 
 | | THE SOLUTION STRUCTURE OF A CYTOTOXIC RIBONUCLEASE FROM THE OOCYTES OF RANA CATESBEIANA (BULLFROG), NMR, 15 STRUCTURES | | Descriptor: | RIBONUCLEASE | | Authors: | Chang, C.-F, Chen, C, Chen, Y.-C, Hom, K, Huang, R.-F, Huang, T. | | Deposit date: | 1998-05-05 | | Release date: | 1998-10-14 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a cytotoxic ribonuclease from the oocytes of Rana catesbeiana (bullfrog).
J.Mol.Biol., 283, 1998
|
|
5XO7
 
 | | Crystal structure of a novel ZEN lactonase mutant with ligand a | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-05-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5XO6
 
 | | Crystal structure of a novel ZEN lactonase mutant | | Descriptor: | PENTAETHYLENE GLYCOL, lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-05-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
2K5K
 
 | | Solution structure of RhR2 from Rhodobacter Sphaeroides. Northeast Structural Genomics Consortium | | Descriptor: | uncharacterized protein RhR2 | | Authors: | Lee, H, Bansal, S, Chen, C.X, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-28 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RhR2
To be Published
|
|
4AUA
 
 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
5X3L
 
 | |
2LA1
 
 | | Expression in Pichia pastoris and backbone dynamics of dendroaspin, a three finger toxin | | Descriptor: | Mambin | | Authors: | Chuang, W.J, Cheng, C.H, Chen, Y.C, Shiu, J.H. | | Deposit date: | 2011-03-01 | | Release date: | 2012-03-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Dynamics and functional differences between dendroaspin and rhodostomin: Insights into protein scaffolds in integrin recognition
Protein Sci., 21, 2012
|
|
8H7P
 
 | | Crystal structure of aqualigase bound with Suc-AAPF | | Descriptor: | 5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, CALCIUM ION, Subtilisin, ... | | Authors: | Li, H, Ma, M.Z, Zhang, L.J, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of aqualigase bound with Suc-AAPF
To Be Published
|
|
6UHX
 
 | | Crystal structure of YIR035C short chain dehydrogenases/reductase from Saccharomyces cerevisiae | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized oxidoreductase YIR035C | | Authors: | Stogios, P.J, Skarina, T, Chen, C, Kagan, O, Iakounine, A, Savchenko, A. | | Deposit date: | 2019-09-29 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of YIR035C short chain dehydrogenases/reductase from Saccharomyces cerevisiae
To Be Published
|
|
1KVZ
 
 | | Solution Structure of Cytotoxic RC-RNase4 | | Descriptor: | RC-RNase4 | | Authors: | Hsu, C.-H, Liao, Y.-D, Chen, L.-W, Wu, S.-H, Chen, C. | | Deposit date: | 2002-01-28 | | Release date: | 2002-07-28 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cytotoxic RNase 4 from the Oocytes of Bullfrog Rana Catesbeiana
J.MOL.BIOL., 326, 2003
|
|
3AXE
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in complex with cellotetraose (cellobiose density was observed) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION, ... | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-04 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
3AXD
 
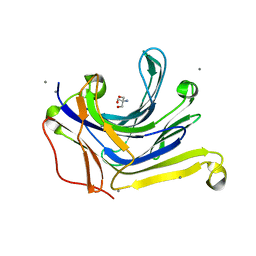 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in apo-form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-03 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
1TI5
 
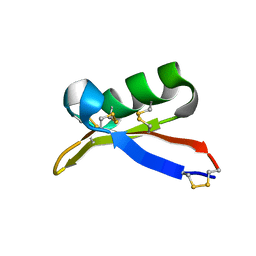 | | Solution structure of plant defensin | | Descriptor: | plant defensin | | Authors: | Liu, Y.J, Cheng, C.S, Liu, Y.N, Hsu, M.P, Chen, C.S, Lyu, P.C. | | Deposit date: | 2004-06-02 | | Release date: | 2005-07-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids.
Proteins, 63, 2006
|
|
2QGU
 
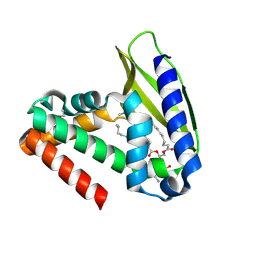 | | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A. Northeast Structural Genomics Consortium target RsR89 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Probable signal peptide protein | | Authors: | Kuzin, A.P, Chen, Y, Jayaraman, S, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A.
To be Published
|
|
2QIK
 
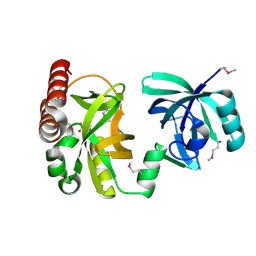 | | Crystal structure of YkqA from Bacillus subtilis. Northeast Structural Genomics Target SR631 | | Descriptor: | CITRIC ACID, UPF0131 protein ykqA | | Authors: | Benach, J, Chen, Y, Forouhar, F, Seetharaman, J, Baran, M.C, Cunningham, K, Ma, L.-C, Owens, L, Chen, C.X, Rong, X, Janjua, H, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of YkqA from Bacillus subtilis.
To be Published
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
4RFU
 
 | | Crystal structure of truncated P-domain from Grouper nervous necrosis virus capsid protein at 1.2A | | Descriptor: | CALCIUM ION, Coat protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
