4XPY
 
 | | Crystal structure of hemerythrin : L114Y mutant | | Descriptor: | Bacteriohemerythrin, FE (II) ION, GLYCEROL | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
4XPX
 
 | | Crystal structure of hemerythrin:wild-type | | Descriptor: | Bacteriohemerythrin, FE (II) ION | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
5IYS
 
 | | Crystal structure of a dehydrosqualene synthase in complex with ligand | | Descriptor: | MAGNESIUM ION, Phytoene synthase, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Liu, G.Z, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of a dehydrosqualene synthase in complex with ligand
to be published
|
|
2O14
 
 | | X-Ray Crystal Structure of Protein YXIM_BACsu from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR595 | | Descriptor: | Hypothetical protein yxiM, MANGANESE (II) ION, SULFATE ION | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray structure of the hypothetical protein YXIM_BACsu from Bacillus subtilis.
To be Published
|
|
2PJF
 
 | | Solution structure of rhodostomin | | Descriptor: | Rhodostoxin-disintegrin rhodostomin | | Authors: | Chuang, W.J, Chen, Y.C, Chen, C.Y, Chang, Y.T. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: Importance of the interactions between the D residue and integrin
Proteins, 2009
|
|
2PJG
 
 | | Solution structure of rhodostomin D51E mutant | | Descriptor: | Rhodostoxin-disintegrin rhodostomin | | Authors: | Chuang, W.J, Chen, Y.C, Chen, C.Y, Chou, L.J. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: Importance of the interactions between the D residue and integrin
Proteins, 2009
|
|
2RQX
 
 | |
5JMT
 
 | | Crystal structure of Zika virus NS3 helicase | | Descriptor: | NS3 helicase | | Authors: | Tian, H, Ji, X, Yang, X, Xie, W, Yang, K, Chen, C, Wu, C, Chi, H, Mu, Z, Wang, Z, Yang, H. | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | The crystal structure of Zika virus helicase: basis for antiviral drug design
Protein Cell, 7, 2016
|
|
2DUA
 
 | |
1XTA
 
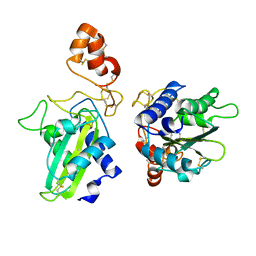 | | Crystal Structure of Natrin, a snake venom CRISP from Taiwan cobra (Naja atra) | | Descriptor: | Natrin 1 | | Authors: | Wang, Y.-L, Goh, K.-X, Lee, S.-C, Huang, W.-N, Wu, W.-G, Chen, C.-J. | | Deposit date: | 2004-10-21 | | Release date: | 2005-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of snake venom CRISP reveal an action mechanism involving serine protease and ion channel blocking domains
To be Published
|
|
8CQI
 
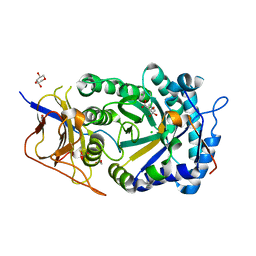 | | Human heparanase in complex with inhibitor R3794 | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 1,5-anhydro-D-arabinitol, ... | | Authors: | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
1JLZ
 
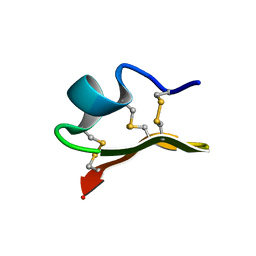 | | Solution Structure of a K+-Channel Blocker from the Scorpion Toxin of Tityus cambridgei | | Descriptor: | Tityustoxin alpha-KTx | | Authors: | Wang, I, Wu, S.-H, Chang, H.-K, Shieh, R.-C, Yu, H.-M, Chen, C. | | Deposit date: | 2001-07-17 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a K(+)-channel blocker from the scorpion Tityus cambridgei.
Protein Sci., 11, 2002
|
|
3E5R
 
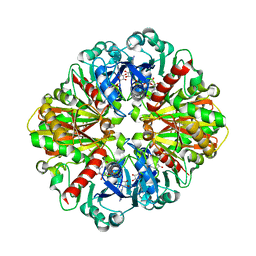 | |
3E6A
 
 | |
4ZXU
 
 | | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289.
To be Published
|
|
1T3S
 
 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, MERCURY (II) ION | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain.
Neuron, 42, 2004
|
|
3DC0
 
 | | Crystal structure of native alpha-amylase from Bacillus sp. KR-8104 | | Descriptor: | CALCIUM ION, alpha-amylase | | Authors: | Alikhajeh, J, Khajeh, K, Ranjbar, B, Naderi-Manesh, M, Naderi-Manesh, H, Chen, C.J. | | Deposit date: | 2008-06-03 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of native alpha-amylase from Bacillus sp. KR-8104
to be published
|
|
3OTW
 
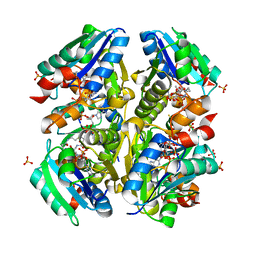 | | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase | | Descriptor: | COENZYME A, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Yin, H.S, Cheng, C.S, Chen, C.G, Luo, Y.C, Chen, W.T, Cheng, S.Y. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase
To be Published
|
|
4LCQ
 
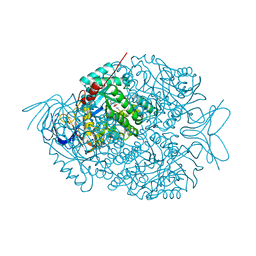 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBI | | Descriptor: | (2S)-3-(carbamoylamino)-2-methylpropanoic acid, ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4LCR
 
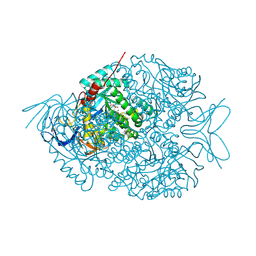 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBA | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
5BQX
 
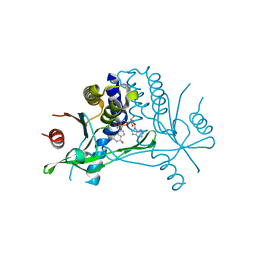 | | Crystal structure of human STING in complex with 3'2'-cGAMP | | Descriptor: | 3'2'-cGAMP, Stimulator of interferon genes protein | | Authors: | Wu, J, Zhang, X, Chen, Z.J, Chen, C. | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2AVF
 
 | | Crystal Structure of C-terminal Desundecapeptide Nitrite Reductase from Achromobacter cycloclastes | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Li, H.T, Chang, T, Chang, W.C, Chen, C.J, Liu, M.Y, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2005-08-30 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of C-terminal desundecapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 338, 2005
|
|
3D23
 
 | | Main protease of HCoV-HKU1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhao, Q, Chen, C, Li, S, Zou, Y. | | Deposit date: | 2008-05-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
J.Virol., 82, 2008
|
|
3ZQC
 
 | | Structure of the Trichomonas vaginalis Myb3 DNA-binding domain bound to a promoter sequence reveals a unique C-terminal beta-hairpin conformation | | Descriptor: | MRE-1, MYB3 | | Authors: | Wei, S.-Y, Lou, Y.-C, Tsai, J.-Y, Hsu, H.-M, Tai, J.-H, Hsiao, C.-D, Chen, C. | | Deposit date: | 2011-06-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Trichomonas Vaginalis Myb3 DNA-Binding Domain Bound to a Promoter Sequence Reveals a Unique C-Terminal Beta-Hairpin Conformation.
Nucleic Acids Res., 40, 2012
|
|
4H00
 
 | | The crystal structure of mon-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
