5JAX
 
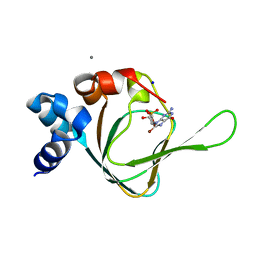 | | PKG I's Carboyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-Br-cGMP | | Descriptor: | 2-amino-8-bromo-9-[(2R,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, SODIUM ION, ... | | Authors: | Campbell, J.C, Sankaran, B, Kim, C.W. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
1EJZ
 
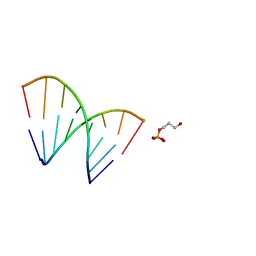 | | SOLUTION STRUCTURE OF A HNA-RNA HYBRID | | Descriptor: | DNA (5'-H(*(6HG)P*(6HC)P*(6HG)P*(6HT)P*(6HA)P*(6HG)P*(6HC)P*(6HG))-3'), PHOSPHORIC ACID MONO-(3-HYDROXY-PROPYL) ESTER, RNA (5'-R(*CP*GP*CP*UP*AP*CP*GP*C)-3') | | Authors: | Lescrnier, E, Esnouf, R, Heus, H.A, Hilbers, C.W, Herdewijn, P. | | Deposit date: | 2000-03-06 | | Release date: | 2000-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a HNA-RNA hybrid.
Chem.Biol., 7, 2000
|
|
2AAD
 
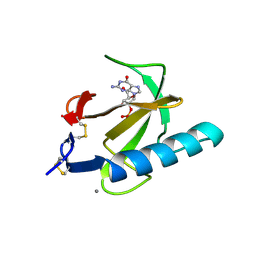 | | THE ROLE OF HISTIDINE-40 IN RIBONUCLEASE T1 CATALYSIS: THREE-DIMENSIONAL STRUCTURES OF THE PARTIALLY ACTIVE HIS40LYS MUTANT | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 ISOZYME | | Authors: | Zegers, I, Verhelst, P, Choe, C.W, Steyaert, J, Heinemann, U, Wyns, L, Saenger, W. | | Deposit date: | 1992-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of histidine-40 in ribonuclease T1 catalysis: three-dimensionalstructures of the partially active His40Lys mutant.
Biochemistry, 31, 1992
|
|
6S7E
 
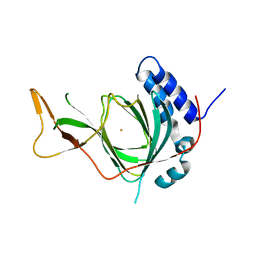 | | Plant Cysteine Oxidase PCO4 from Arabidopsis thaliana (using PEG 3350 and NaF as precipitants) | | Descriptor: | FE (III) ION, Plant cysteine oxidase 4 | | Authors: | White, M.D, Levy, C.W, Flashman, E, McDonough, M.A. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of Arabidopsis thaliana oxygen-sensing plant cysteine oxidases 4 and 5 enable targeted manipulation of their activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2OTO
 
 | |
5C5T
 
 | |
5C5U
 
 | | The crystal structure of viral collagen prolyl hydroxylase vCPH from Paramecium Bursaria Chlorella virus-1 - Truncated Construct | | Descriptor: | ACETATE ION, MANGANESE (II) ION, Prolyl 4-hydroxylase, ... | | Authors: | Longbotham, J.E, Levy, C.W, Johannisen, L.O, Tarhonskaya, H, Jiang, S, Loenarz, C, Flashman, E, Hay, S, Schofiled, C.J, Scrutton, N.S. | | Deposit date: | 2015-06-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Mechanism of a Viral Collagen Prolyl Hydroxylase.
Biochemistry, 54, 2015
|
|
5C6C
 
 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cAMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CADMIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-22 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
5C8W
 
 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MALONIC ACID, SODIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
1KOZ
 
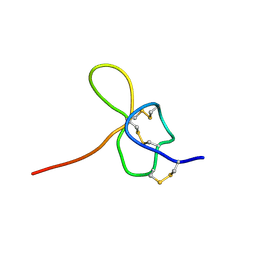 | | SOLUTION STRUCTURE OF OMEGA-GRAMMOTOXIN SIA | | Descriptor: | Voltage-dependent Channel Inhibitor | | Authors: | Takeuchi, K, Park, E.J, Lee, C.W, Kim, J.I, Takahashi, H, Swartz, K.J, Shimada, I. | | Deposit date: | 2001-12-25 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega-grammotoxin SIA, a gating modifier of P/Q and N-type Ca(2+) channel.
J.Mol.Biol., 321, 2002
|
|
5GPG
 
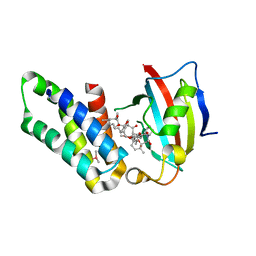 | | Co-crystal structure of the FK506 binding domain of human FKBP25, Rapamycin and the FRB domain of human mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP3, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Lee, H.B, Lee, S.Y, Rhee, H.W, Lee, C.W. | | Deposit date: | 2016-08-02 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proximity-Directed Labeling Reveals a New Rapamycin-Induced Heterodimer of FKBP25 and FRB in Live Cells
Acs Cent.Sci., 2, 2016
|
|
7O1D
 
 | |
4O1Q
 
 | |
6Q7P
 
 | | Crystal structure of OE1.2 | | Descriptor: | 1,2-ETHANEDIOL, 1-PHENYLETHANONE, MAGNESIUM ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6Q7R
 
 | |
6Q7O
 
 | | Crystal structure of OE1 | | Descriptor: | CALCIUM ION, OE1 | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6Q7N
 
 | |
6Q7Q
 
 | | Crystal structure of OE1.3 | | Descriptor: | OE1.3 | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
7JIH
 
 | | HRAS A59E GppNHp | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
6GIA
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant I107A | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6GI7
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant L25I | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6GKV
 
 | | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | | Descriptor: | 6,7-dimethoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-2-ium, Coclaurine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Dunstan, M.S, Levy, C.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Biocatalytic Scope of Coclaurine N-Methyltransferase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GI8
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant L25A | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6GI9
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant I107L | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6GKY
 
 | |
