6W91
 
 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | Nonstructural polyprotein, methyl 2-oxo-2,5-dihydropyrimidine-4-carboxylate | | Authors: | Zhang, S, Garzan, A, Pathak, A.K, Augelli-Szafran, C.E, Wu, M. | | Deposit date: | 2020-03-21 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6V3A
 
 | |
2WWN
 
 | | Yersinia pseudotuberculosis Superoxide Dismutase C with bound Azide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AZIDE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Basak, A.K, Duffield, M.L, Naylor, C.E, Huyet, J, Titball, R.W. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Yersinia Pseudotuberculosis Superoxide Dismutase (Sodc)
To be Published
|
|
6V3D
 
 | |
2YGT
 
 | | Clostridium perfringens delta-toxin | | Descriptor: | DELTA TOXIN, GLYCEROL, IMIDAZOLE, ... | | Authors: | Huyet, J, Naylor, C.E, Gibert, M, Popoff, M.R, Basak, A.K. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Clostridium Perfringens Delta Toxin Pore Formation.
Plos One, 8, 2013
|
|
2YHJ
 
 | | Clostridium perfringens Enterotoxin at 4.0 Angstrom Resolution | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, McClane, B.A, Basak, A.K. | | Deposit date: | 2011-05-03 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
2Y3E
 
 | | Traptavidin, apo-form | | Descriptor: | GLYCEROL, STREPTAVIDIN | | Authors: | Chivers, C.E, Koner, A.L, Lowe, E.D, Howarth, M. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | How the Biotin-Streptavidin Interaction Was Made Even Stronger: Investigation Via Crystallography and a Chimeric Tetramer.
Biochem.J., 435, 2011
|
|
5A6P
 
 | | Heavy metal associated domain of NLR-type immune receptor Pikp1 from rice (Oryza sativa) | | Descriptor: | RESISTANCE PROTEIN PIKP-1 | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
1BJT
 
 | | TOPOISOMERASE II RESIDUES 409-1201 | | Descriptor: | TOPOISOMERASE II | | Authors: | Fass, D, Bogden, C.E, Berger, J.M. | | Deposit date: | 1998-06-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands.
Nat.Struct.Biol., 6, 1999
|
|
1BIW
 
 | | DESIGN AND SYNTHESIS OF CONFORMATIONALLY-CONSTRAINED MMP INHIBITORS | | Descriptor: | CALCIUM ION, N1-HYDROXY-2-(3-HYDROXY-PROPYL)-3-ISOBUTYL-N4-[1-(2-METHOXY-ETHYL)-2-OXO-AZEPAN-3-YL]-SUCCINAMIDE, PROTEIN (STROMELYSIN-1 COMPLEX), ... | | Authors: | Natchus, M.G, Cheng, M, Wahl, C.T, Pikul, S, Almstead, N.G, Bradley, R.S, Taiwo, Y.O, Mieling, G.E, Dunaway, C.M, Snider, C.E, McIver, J.M, Barnett, B.L, McPhail, S.J, Anastasio, M.B, De, B. | | Deposit date: | 1998-06-19 | | Release date: | 1999-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of conformationally-constrained MMP inhibitors.
Bioorg.Med.Chem.Lett., 8, 1998
|
|
1BO5
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN ESCHERICHIA COLI GLYCEROL KINASE AND THE ALLOSTERIC REGULATOR FRUCTOSE 1,6-BISPHOSPHATE. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, PROTEIN (GLYCEROL KINASE) | | Authors: | Ormo, M, Bystrom, C.E, Remington, S.J. | | Deposit date: | 1998-08-10 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate.
Biochemistry, 37, 1998
|
|
1BOT
 
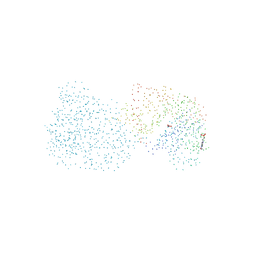 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN ESCHERICHIA COLI GLYCEROL KINASE AND THE ALLOSTERIC REGULATOR FRUCTOSE 1,6-BISPHOSPHATE. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (GLYCEROL KINASE) | | Authors: | Ormo, M, Bystrom, C.E, Remington, S.J. | | Deposit date: | 1998-08-05 | | Release date: | 1999-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate.
Biochemistry, 37, 1998
|
|
1BTS
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-11-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
5BRW
 
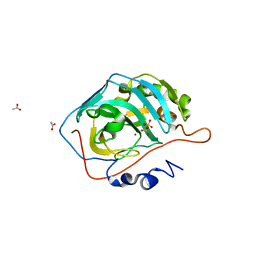 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
1D1J
 
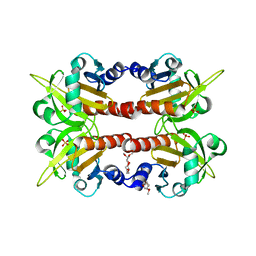 | | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, PROFILIN II, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Lindberg, U, Schutt, C.E. | | Deposit date: | 1999-09-17 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure determination of human profilin II: A comparative structural analysis of human profilins.
J.Mol.Biol., 294, 1999
|
|
1E3Q
 
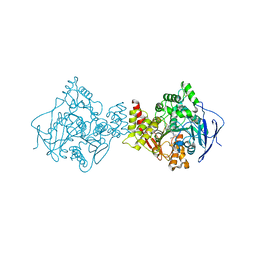 | | TORPEDO CALIFORNICA ACETYLCHOLINESTERASE COMPLEXED WITH BW284C51 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(5-{4-[DIMETHYL(PROP-2-ENYL)AMMONIO]PHENYL}-3-OXOPENTYL)-N,N-DIMETHYL-N-PROP-2-ENYLBENZENAMINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Felder, C.E, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2000-06-21 | | Release date: | 2000-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of a Complex of the Potent and Specific Inhibitor Bw284C51 with Torpedo Californica Acetylcholinesterase
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5CEE
 
 | | Malic enzyme from Candidatus Phytoplasma AYWB in complex with NAD and Mg2+ | | Descriptor: | MAGNESIUM ION, NAD-dependent malic enzyme, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Alvarez, C.E, Trajtenberg, F, Larrieux, N, Saigo, M, Mussi, M.A, Andreo, C.S, Drincovich, M.F, Buschiazzo, A. | | Deposit date: | 2015-07-06 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | The crystal structure of the malic enzyme from Candidatus Phytoplasma reveals the minimal structural determinants for a malic enzyme.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5CPL
 
 | | The crystal structure of Xenobiotic reductase A (XenA) from Pseudomonas putida in complex with a nicotinamide mimic (mNH2) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 1-benzyl-1,4,5,6-tetrahydropyridine-3-carboxamide, CALCIUM ION, ... | | Authors: | Knaus, T, Paul, C.E, Levy, C.W, Mutti, F.G, Hollmann, F, Scrutton, N.S. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5CPM
 
 | | XenA from Pseudomonas putida in complex with NADPH4. | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Xenobiotic reductase | | Authors: | Knaus, T, Paul, C.E, Levy, C.W, Mutti, F.G, Hollmann, F, Scrutton, N.S. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5CPN
 
 | | Crystal structure of XenA from Pseudomonas putida in complex with an NADH mimic (mAc) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 1-[(3S)-1-benzylpiperidin-3-yl]ethanone, Xenobiotic reductase | | Authors: | Knaus, T, Paul, C.E, Levy, C.W, Mutti, F.G, Hollmann, F, Scrutton, N.S. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5A15
 
 | | Crystal structure of the BTB domain of human KCTD16 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD16 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Canning, P, Dixon Clarke, S.E, Talon, R, Wiggers, H.J, Fitzpatrick, F, Tallant, C, Kopec, J, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-04-28 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
5A6W
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with the HMA domain of Pikp1 from rice (Oryza sativa) | | Descriptor: | 1,2-ETHANEDIOL, AVR-PIK PROTEIN, RESISTANCE PROTEIN PIKP-1, ... | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-07-01 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
5A76
 
 | |
1E08
 
 | | Structural model of the [Fe]-Hydrogenase/cytochrome c553 complex combining NMR and soft-docking | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Morelli, X, Czjzek, M, Hatchikian, C.E, Bornet, O, Fontecilla-Camps, J.C, Palma, N.P, Moura, J.J.G, Guerlesquin, F. | | Deposit date: | 2000-03-13 | | Release date: | 2000-08-25 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Fe-Hydrogenase/Cytochrome C553 Complex Combining Transverse Relaxation-Optimized Spectroscopy Experiments and Soft Docking Calculations.
J.Biol.Chem., 275, 2000
|
|
1BTQ
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-11-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
