1RIY
 
 | |
1UJL
 
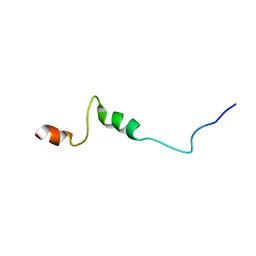 | | Solution Structure of the HERG K+ channel S5-P extracellular linker | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Torres, A.M, Bansal, P.S, Sunde, M, Clarke, C.E, Bursill, J.A, Smith, D.J, Bauskin, A, Breit, S.N, Campbell, T.J, Alewood, P.F, Kuchel, P.W, Vandenberg, J.I. | | Deposit date: | 2003-08-05 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the HERG K+ channel S5P extracellular linker: role of an amphipathic alpha-helix
in C-type inactivation.
J.Biol.Chem., 278, 2003
|
|
7SSW
 
 | |
7SSN
 
 | |
7SS9
 
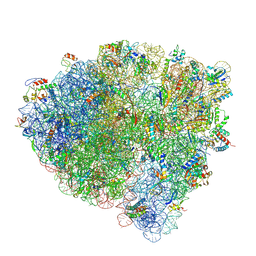 | | Late translocation intermediate with EF-G partially dissociated (Structure V) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Carbone, C.E, Loveland, A.B, Gamper, H.B, Hou, Y, Korostelev, A.A. | | Deposit date: | 2021-11-10 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP.
Nat Commun, 12, 2021
|
|
7UXX
 
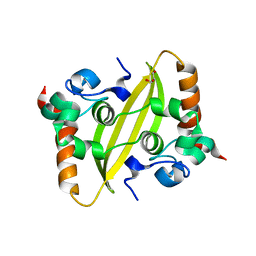 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleoprotein | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
7UXZ
 
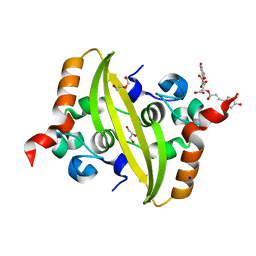 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain complexed with Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
1SDQ
 
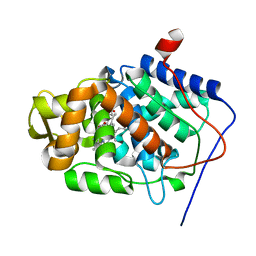 | | Structure of reduced-NO adduct of mesopone cytochrome c peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, FE-(4-MESOPORPHYRINONE)-R-ISOMER, ... | | Authors: | Bhaskar, B, Immoos, C.E, Sulc, F, Cohem, M.S, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2004-02-13 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structures of resting (Fe3+), reduced (Fe2+) and NO-bound states of mesopone cytochrome c peroxidase (MpCcP) (R-isomer)
To be Published
|
|
1SG6
 
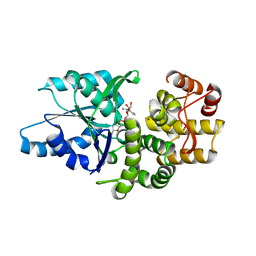 | | Crystal structure of Aspergillus nidulans 3-dehydroquinate synthase (AnDHQS) in complex with Zn2+ and NAD+, at 1.7D | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pentafunctional AROM polypeptide, ZINC ION | | Authors: | Nichols, C.E, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the 'open' form of Aspergillus nidulans 3-dehydroquinate synthase at 1.7 A resolution from crystals grown following enzyme turnover.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T0C
 
 | |
1USL
 
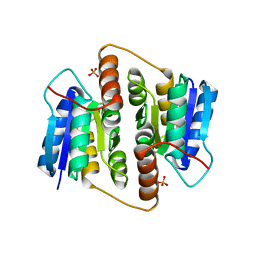 | | Structure Of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, Complexed With Phosphate. | | Descriptor: | PHOSPHATE ION, RIBOSE 5-PHOSPHATE ISOMERASE B | | Authors: | Roos, A.K, Andersson, C.E, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-11-25 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase Has a Known Fold, But a Novel Active Site
J.Mol.Biol., 335, 2004
|
|
1V1P
 
 | |
1VCB
 
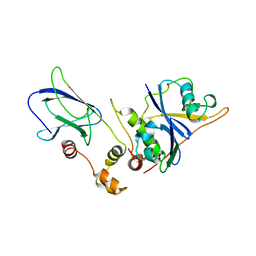 | | THE VHL-ELONGINC-ELONGINB STRUCTURE | | Descriptor: | PROTEIN (ELONGIN B), PROTEIN (ELONGIN C), PROTEIN (VHL) | | Authors: | Stebbins, C.E, Kaelin, W.G, Pavletich, N.P. | | Deposit date: | 1999-03-13 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function.
Science, 284, 1999
|
|
1SFL
 
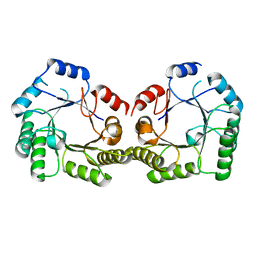 | | 1.9A Crystal structure of Staphylococcus aureus type I 3-dehydroquinase, apo form | | Descriptor: | 3-dehydroquinate dehydratase | | Authors: | Nichols, C.E, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-20 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Staphylococcus aureus type I dehydroquinase from enzyme turnover experiments.
Proteins, 56, 2004
|
|
1SR4
 
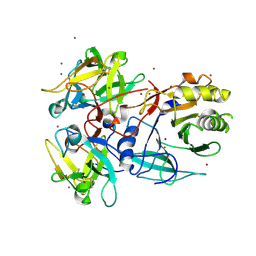 | | Crystal Structure of the Haemophilus ducreyi cytolethal distending toxin | | Descriptor: | BROMIDE ION, Cytolethal distending toxin subunit A, cytolethal distending toxin protein B, ... | | Authors: | Nesic, D, Hsu, Y, Stebbins, C.E. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Assembly and Function of a Bacterial Genotoxin
Nature, 429, 2004
|
|
1UZU
 
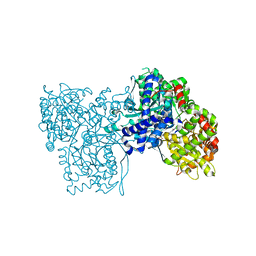 | | Glycogen Phosphorylase b in complex with indirubin-5'-sulphonate | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Kosmopoulou, M.N, Leonidas, D.D, Chrysina, E.D, Bischler, N, Eisenbrand, G, Sakarellos, C.E, Pauptit, R, Oikonomakos, N.G. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of the potential antitumour agent indirubin-5-sulphonate at the inhibitor site of rabbit muscle glycogen phosphorylase b. Comparison with ligand binding to pCDK2-cyclin A complex.
Eur. J. Biochem., 271, 2004
|
|
1V1O
 
 | |
1URF
 
 | | HR1b domain from PRK1 | | Descriptor: | PROTEIN KINASE C-LIKE 1 | | Authors: | Owen, D, Lowe, P.N, Nietlispach, D, Brosnan, C.E, Chirgadze, D.Y, Parker, P.J, Blundell, T.L, Mott, H.R. | | Deposit date: | 2003-10-29 | | Release date: | 2003-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Dissection of the Interaction between the Small G Proteins Rac1 and Rhoa and Protein Kinase C-Related Kinase 1 (Prk1)
J.Biol.Chem., 278, 2003
|
|
5A6P
 
 | | Heavy metal associated domain of NLR-type immune receptor Pikp1 from rice (Oryza sativa) | | Descriptor: | RESISTANCE PROTEIN PIKP-1 | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
5A15
 
 | | Crystal structure of the BTB domain of human KCTD16 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD16 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Canning, P, Dixon Clarke, S.E, Talon, R, Wiggers, H.J, Fitzpatrick, F, Tallant, C, Kopec, J, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-04-28 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
5A6W
 
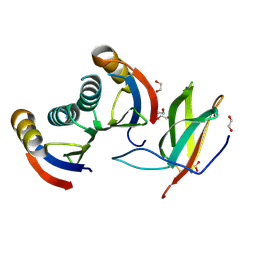 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with the HMA domain of Pikp1 from rice (Oryza sativa) | | Descriptor: | 1,2-ETHANEDIOL, AVR-PIK PROTEIN, RESISTANCE PROTEIN PIKP-1, ... | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-07-01 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
5A76
 
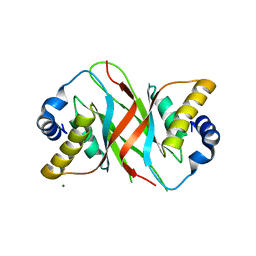 | |
5A6R
 
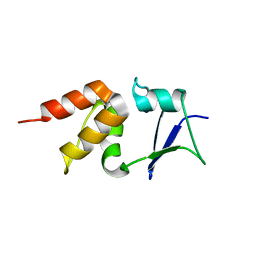 | | Crystal structure of the BTB domain of human KCTD17 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD17 | | Authors: | Pinkas, D.M, Sorrell, F, Sanvitale, C.E, Goubin, S, Williams, E, Newman, J.A, Pearce, N.M, Neshich, I, Pike, A.C.W, MacKenzie, A, Quigley, A, Faust, B, Carpenter, E.P, Tallant, C, Kopec, J, Chalk, R, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
5CEE
 
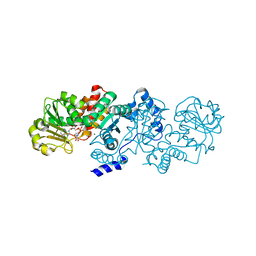 | | Malic enzyme from Candidatus Phytoplasma AYWB in complex with NAD and Mg2+ | | Descriptor: | MAGNESIUM ION, NAD-dependent malic enzyme, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Alvarez, C.E, Trajtenberg, F, Larrieux, N, Saigo, M, Mussi, M.A, Andreo, C.S, Drincovich, M.F, Buschiazzo, A. | | Deposit date: | 2015-07-06 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | The crystal structure of the malic enzyme from Candidatus Phytoplasma reveals the minimal structural determinants for a malic enzyme.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4ZRT
 
 | | PTP1BC215S bound to Nephrin peptide substrate | | Descriptor: | CHLORIDE ION, GLY-PRO-LEU-PTR-ASP-GLU, GLYCEROL, ... | | Authors: | Selner, N.G, Bell, C.E, Pei, D. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-24 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Diverse levels of sequence selectivity and catalytic efficiency of protein-tyrosine phosphatases.
Biochemistry, 53, 2014
|
|
