3R5L
 
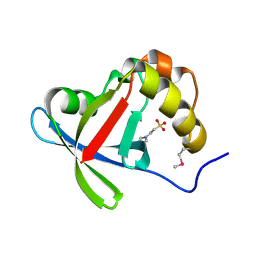 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5R
 
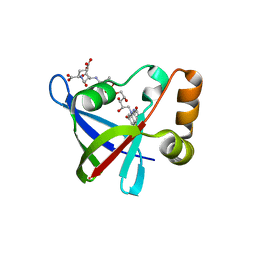 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3MTH
 
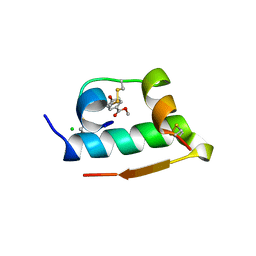 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, CHLORIDE ION, METHYLPARABEN INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
3R99
 
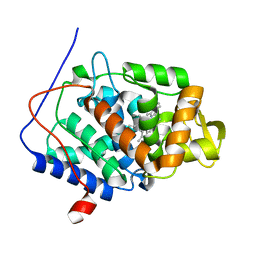 | | Joint Neutron and X-ray structure of Cytochrome c peroxidase | | Descriptor: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Blakeley, M.P, Fisher, S.J, Gumiero, A, Moody, P.C.E, Raven, E.L. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (2.4 Å), X-RAY DIFFRACTION | | Cite: | Hydrogen bonds in heme peroxidases: a combined X-ray and neutron study of cytochrome c peroxidase
To be Published
|
|
3R5Z
 
 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
2FM8
 
 | |
3R5P
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | Deazaflavin-dependent nitroreductase, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3RLL
 
 | | Crystal structure of the T877A androgen receptor ligand binding domain in complex with (S)-N-(4-Cyano-3-(trifluoromethyl)phenyl)-3-(4-cyanonaphthalen-1-yloxy)-2-hydroxy-2-methylpropanamide | | Descriptor: | (2S)-3-[(4-cyanonaphthalen-1-yl)oxy]-N-[4-cyano-3-(trifluoromethyl)phenyl]-2-hydroxy-2-methylpropanamide, Androgen receptor | | Authors: | Bohl, C.E, Duke, C.B, Jones, A, Dalton, J.T, Miller, D.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected binding orientation of bulky-B-ring anti-androgens and implications for future drug targets.
J.Med.Chem., 54, 2011
|
|
3R5W
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R98
 
 | | Joint Neutron and X-ray structure of Cytochrome c peroxidase | | Descriptor: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Blakeley, M.P, Fisher, S.J, Gumiero, A, Moody, P.C.E, Raven, E.L. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (2.4 Å), X-RAY DIFFRACTION | | Cite: | Hydrogen bonds in heme peroxidases: a combined X-ray and neutron study of cytochrome c peroxidase
To be Published
|
|
6Y8N
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y80
 
 | | Fragment KCL_916 in complex with MAP kinase p38-alpha | | Descriptor: | 1-(2-adamantylmethyl)-3-ethyl-guanidine, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6Y81
 
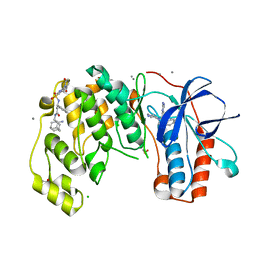 | | Fragment KCL_1088 in complex with MAP kinase p38-alpha | | Descriptor: | (3~{R})-~{N}-[(2-azanyl-2-adamantyl)methyl]-3-[[6-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]pyridin-3-yl]sulfonylamino]-3-phenyl-propanamide, 1,2-ETHANEDIOL, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6YO1
 
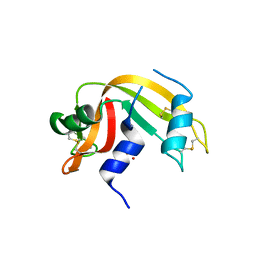 | | Crystal structure of ribonuclease A solved by vanadium SAD phasing | | Descriptor: | Ribonuclease pancreatic, URIDINE-2',3'-VANADATE | | Authors: | El Omari, K, Mohamad, N, Bountra, K, Duman, R, Romano, M, Schlegel, K, Kwong, H, Mykhaylyk, V, Olesen, C.E, Moller, J.V, Bublitz, M, Beis, K, Wagner, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental phasing with vanadium and application to nucleotide-binding membrane proteins.
Iucrj, 7, 2020
|
|
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6Y82
 
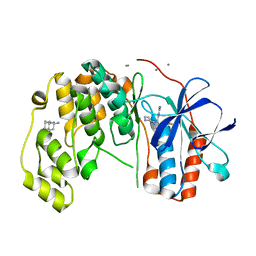 | | Fragment KCL_804 in complex with MAP kinase p38-alpha | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Fragment KCL_804 in complex with MAP kinase p38-alpha
To Be Published
|
|
6Y8H
 
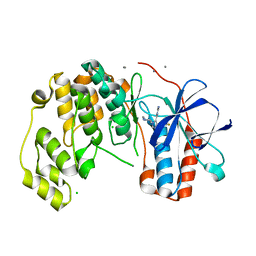 | |
6YCW
 
 | | Fragment KCL_K767 in complex with MAP kinase p38-alpha | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, Mitogen-activated protein kinase 14, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-19 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6Y7X
 
 | | Fragment KCL_771 in complex with MAP kinase p38-alpha | | Descriptor: | (2-azanyl-2-adamantyl)methanol, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
4YFX
 
 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
6Y8M
 
 | |
6YCU
 
 | | Fragment KCL_K777 in complex with MAP kinase p38-alpha | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, 4-[2,5-bis(oxidanylidene)pyrrol-1-yl]-~{N}-propyl-benzenesulfonamide, CALCIUM ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-19 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6Y7W
 
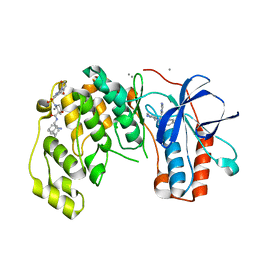 | | Fragment KCL_1337 in complex with MAP kinase p38-alpha | | Descriptor: | (2~{R})-~{N}-[(2-azanyl-2-adamantyl)methyl]-4-[6-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]pyridin-3-yl]sulfonyl-2-methyl-morpholine-2-carboxamide, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
4YFN
 
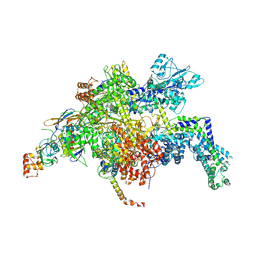 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
