2C74
 
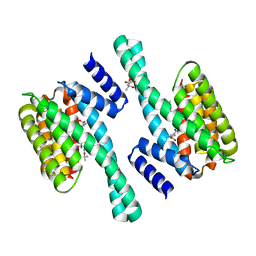 | | 14-3-3 Protein Eta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN ETA, CITRIC ACID, CONSENSUS PEPTIDE MODE 1 FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Yang, X, Smee, C.E.A, Johansson, C, Sundstrom, M, Edwards, A, Weigelt, J, Arrowsmith, C, Doyle, D.A. | | Deposit date: | 2005-11-17 | | Release date: | 2005-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for protein-protein interactions in the 14-3-3 protein family.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
1LM3
 
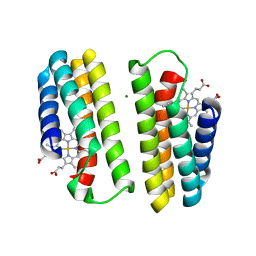 | | A Multi-generation Analysis of Cytochrome b562 Redox Variants: Evolutionary Strategies for Modulating Redox Potential Revealed Using a Library Approach | | Descriptor: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, SOLUBLE CYTOCHROME B562 | | Authors: | Springs, S.L, Bass, S.E, Bowman, G, Nodelman, I, Schutt, C.E, McLendon, G.L. | | Deposit date: | 2002-04-30 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A multigeneration analysis of cytochrome b(562) redox variants: evolutionary strategies for modulating redox potential revealed using a library approach.
Biochemistry, 41, 2002
|
|
5A6P
 
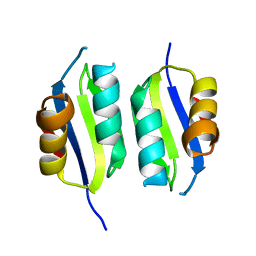 | | Heavy metal associated domain of NLR-type immune receptor Pikp1 from rice (Oryza sativa) | | Descriptor: | RESISTANCE PROTEIN PIKP-1 | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
5BRW
 
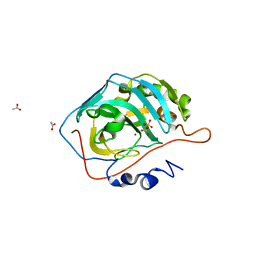 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
5ELP
 
 | |
2CJJ
 
 | | Crystal Structure of the MYB domain of the RAD transcription factor from Antirrhinum majus | | Descriptor: | RADIALIS | | Authors: | Stevenson, C.E.M, Burton, N, Costa, M.M, Nath, U, Dixon, R.A, Coen, E.S, Lawson, D.M. | | Deposit date: | 2006-04-04 | | Release date: | 2006-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Myb Domain of the Rad Transcription Factor from Antirrhinum Majus.
Proteins: Struct., Funct., Bioinf., 65, 2006
|
|
1LW0
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
5EKA
 
 | | HU DNA-binding protein from Thermus thermophilus | | Descriptor: | DNA-binding protein HU, GLYCEROL | | Authors: | Papageorgiou, A, Adam, P, Stavros, P, Nounesis, G, Meijers, R, Petratos, K, Vorgias, C.E. | | Deposit date: | 2015-11-03 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | HU histone-like DNA-binding protein from Thermus thermophilus: structural and evolutionary analyses.
Extremophiles, 20, 2016
|
|
5EL2
 
 | |
5F6F
 
 | |
1NVF
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+, ADP and carbaphosphonate | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
1NVE
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and NAD | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
2CL4
 
 | | Ascorbate Peroxidase R172A mutant | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Ghamsari, L, MacDonald, I.K, Moody, P.C.E. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of Ascorbate Peroxidase with Substrates: A Mechanistic and Structural Analysis.
Biochemistry, 45, 2006
|
|
5F91
 
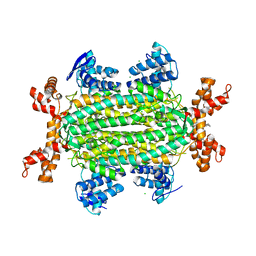 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator (N-(5-(azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fumarate hydratase class II, ... | | Authors: | Kasbekar, M, Fischer, G, Mott, B.T, Yasgar, A, Hyvonen, M, Boshoff, H.I, Abell, C, Barry, C.E, Thomas, C.J. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Selective small molecule inhibitor of the Mycobacterium tuberculosis fumarate hydratase reveals an allosteric regulatory site.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1OAF
 
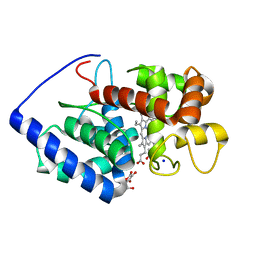 | | Ascobate peroxidase from soybean cytosol in complex with ascorbate | | Descriptor: | ASCORBATE PEROXIDASE, ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2003-01-13 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Ascorbate Peroxidase-Ascorbate Complex
Nat.Struct.Biol., 10, 2003
|
|
1OAG
 
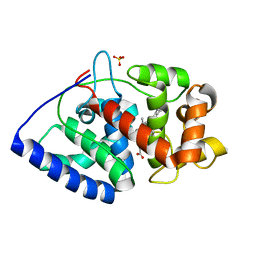 | |
2AWY
 
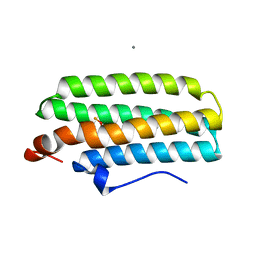 | | met-DcrH-Hr | | Descriptor: | CALCIUM ION, CHLORIDE ION, MU-OXO-DIIRON, ... | | Authors: | Isaza, C.E, Silaghi-Dumitrescu, R, Iyer, R.B, Kurtz, D.M, Chan, M.K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for O(2) Sensing by the Hemerythrin-like Domain of a Bacterial Chemotaxis Protein: Substrate Tunnel and Fluxional N Terminus
Biochemistry, 45, 2006
|
|
9B8O
 
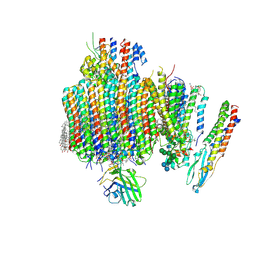 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, Vo | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles
Science, 2024
|
|
2OPP
 
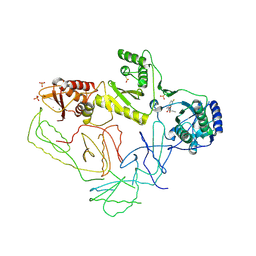 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Chan, S.J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
1AAJ
 
 | |
1BJT
 
 | | TOPOISOMERASE II RESIDUES 409-1201 | | Descriptor: | TOPOISOMERASE II | | Authors: | Fass, D, Bogden, C.E, Berger, J.M. | | Deposit date: | 1998-06-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands.
Nat.Struct.Biol., 6, 1999
|
|
1BIW
 
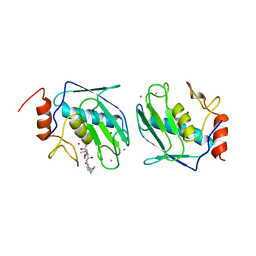 | | DESIGN AND SYNTHESIS OF CONFORMATIONALLY-CONSTRAINED MMP INHIBITORS | | Descriptor: | CALCIUM ION, N1-HYDROXY-2-(3-HYDROXY-PROPYL)-3-ISOBUTYL-N4-[1-(2-METHOXY-ETHYL)-2-OXO-AZEPAN-3-YL]-SUCCINAMIDE, PROTEIN (STROMELYSIN-1 COMPLEX), ... | | Authors: | Natchus, M.G, Cheng, M, Wahl, C.T, Pikul, S, Almstead, N.G, Bradley, R.S, Taiwo, Y.O, Mieling, G.E, Dunaway, C.M, Snider, C.E, McIver, J.M, Barnett, B.L, McPhail, S.J, Anastasio, M.B, De, B. | | Deposit date: | 1998-06-19 | | Release date: | 1999-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of conformationally-constrained MMP inhibitors.
Bioorg.Med.Chem.Lett., 8, 1998
|
|
1BO5
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN ESCHERICHIA COLI GLYCEROL KINASE AND THE ALLOSTERIC REGULATOR FRUCTOSE 1,6-BISPHOSPHATE. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, PROTEIN (GLYCEROL KINASE) | | Authors: | Ormo, M, Bystrom, C.E, Remington, S.J. | | Deposit date: | 1998-08-10 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate.
Biochemistry, 37, 1998
|
|
