4XGW
 
 | | Crystal structure of Escherichia coli Flavin trafficking protein, an FMN transferase, E169K mutant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
4XXO
 
 | |
4XRR
 
 | | Crystal structure of cals8 from micromonospora echinospora (P294S mutant) | | Descriptor: | CalS8, GLYCEROL | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.
J. Biol. Chem., 290, 2015
|
|
4XT0
 
 | | Crystal Structure of Beta-etherase LigF from Sphingobium sp. strain SYK-6 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, PENTAETHYLENE GLYCOL, ... | | Authors: | Helmich, K.E, Bingman, C.A, Donohue, T.J, Phillips Jr, G.N. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of beta-Aryl Ether Bonds in Lignin.
J.Biol.Chem., 291, 2016
|
|
4XUJ
 
 | |
3SNM
 
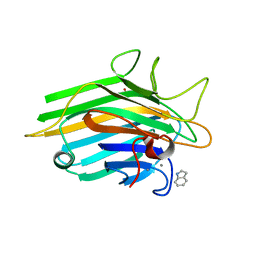 | | Crystal structure of a lectin from Canavalia maritima seeds complexed with Indole-3-Acetic Acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, CALCIUM ION, Concanavalin-A, ... | | Authors: | Delatorre, P, Silva-Filho, J.C, Nobrega, R.B, Rocha, B.C, Cavada, B.S, Gadelha, C.A.A, Santi-Gadelha, T, Alencar, K.L. | | Deposit date: | 2011-06-29 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interactions between indole-3-acetic acid (IAA) with a lectin from Canavalia maritima seeds reveal a new function for lectins in plant physiology.
Biochimie, 95, 2013
|
|
7RRM
 
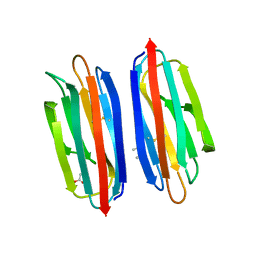 | | Structure of the human TMED1 (p24gamma1) Golgi dynamics Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Transmembrane emp24 domain-containing protein 1 | | Authors: | Mota, D.C.A.M, Cardoso, I.A, Mori, R.M, Mendes, L.F.S, Nonato, M.C, Filho, A.J.C. | | Deposit date: | 2021-08-10 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and thermodynamic analyses of human TMED1 (p24 gamma 1) Golgi dynamics.
Biochimie, 192, 2022
|
|
7L5X
 
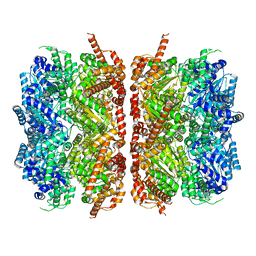 | | p97-R155H mutant dodecamer II | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7L5W
 
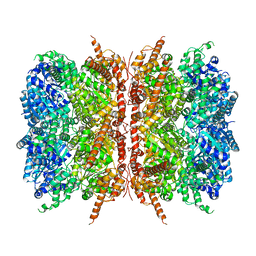 | | p97-R155H mutant dodecamer I | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
5R42
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 7.5, room temperature | | Descriptor: | IODIDE ION, gamma-Chymotrypsin, peptide SWPW, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R49
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 5.6, cryo temperature | | Descriptor: | IODIDE ION, MALONATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R45
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 7.5, cryo temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, MALONATE ION, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4C
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 9, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R48
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 5.6, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4A
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 9, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R44
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 7.5, room temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, peptide SWPW, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R46
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 5.6, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R43
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 7.5, cryo temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, MALONIC ACID, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4B
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 9, cryo temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4D
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 9, cryo temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R47
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 5.6, cryo temperature | | Descriptor: | IODIDE ION, MALONIC ACID, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
7TQR
 
 | |
7JU2
 
 | | Crystal structure of the monomeric ETV6 PNT domain | | Descriptor: | FORMIC ACID, Transcription factor ETV6 | | Authors: | Gerak, C.A.N, Kolesnikov, M, Murphy, M.E.P, McIntosh, L.P. | | Deposit date: | 2020-08-19 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85002184 Å) | | Cite: | Biophysical characterization of the ETV6 PNT domain polymerization interfaces.
J.Biol.Chem., 296, 2021
|
|
7TLX
 
 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
1Y7N
 
 | | Solution structure of the second PDZ domain of the human neuronal adaptor X11alpha | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 1 | | Authors: | Duquesne, A.E, de Ruijter, M, Brouwer, J, Drijfhout, J.W, Nabuurs, S.B, Spronk, C.A.E.M, Vuister, G.W, Ubbink, M, Canters, G.W. | | Deposit date: | 2004-12-09 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of the neuronal adaptor X11alpha and its interaction with the C-terminal peptide of the human copper chaperone for superoxide dismutase
J.Biomol.Nmr, 32, 2005
|
|
