4EQJ
 
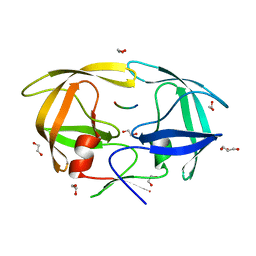 | | Crystal Structure of inactive single chain variant of HIV-1 Protease in Complex with the substrate RT-RH | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
1NP4
 
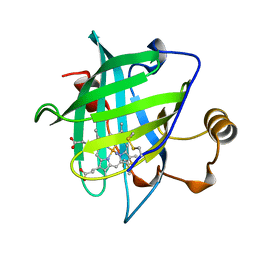 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, PROTEIN (NITROPHORIN 4), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Andersen, J.F, Weichsel, A, Champagne, D.E, Balfour, C.A, Montfort, W.R. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of nitrophorin 4 at 1.5 A resolution: transport of nitric oxide by a lipocalin-based heme protein.
Structure, 6, 1998
|
|
2OYE
 
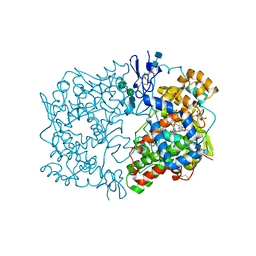 | | Indomethacin-(R)-alpha-ethyl-ethanolamide bound to Cyclooxygenase-1 | | Descriptor: | 2-[1-(4-CHLOROBENZOYL)-5-METHOXY-2-METHYL-1H-INDOL-3-YL]-N-[(1R)-1-(HYDROXYMETHYL)PROPYL]ACETAMIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harman, C.A, Garavito, R.M. | | Deposit date: | 2007-02-21 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of enantioselective inhibition of cyclooxygenase-1 by S-alpha-substituted indomethacin ethanolamides.
J.Biol.Chem., 282, 2007
|
|
4LRB
 
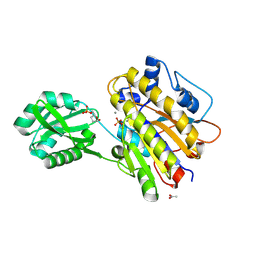 | | Phosphopentomutase S154G variant soaked with 2,3-dideoxyribose 5-phosphate | | Descriptor: | 2,3-dideoxy-5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, GLYCEROL, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
3U65
 
 | | The Crystal Structure of Tat-P(T) (Tp0957) | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, Tp33 protein | | Authors: | Brautigam, C.A, Tomchick, D.R, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
3NJO
 
 | | X-ray crystal structure of the Pyr1-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYR1, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Peterson, F.C, Volkman, B.F, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4LR8
 
 | | Phosphopentomutase S154A variant soaked with ribose 5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
2GU2
 
 | | Crystal Structure of an Aspartoacylase from Rattus norvegicus | | Descriptor: | Aspa protein, SULFATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-06-20 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3TVT
 
 | |
1NND
 
 | | Arginine 116 is Essential for Nucleic Acid Recognition by the Fingers Domain of Moloney Murine Leukemia Virus Reverse Transcriptase | | Descriptor: | Reverse Transcriptase | | Authors: | Crowther, R.L, Remeta, D.P, Minetti, C.A, Das, D, Montano, S.P, Georgiadis, M.M. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and energetic characterization of nucleic acid-binding to the fingers domain of Moloney murine leukemia virus reverse transcriptase
Proteins, 57, 2004
|
|
4E4D
 
 | | Crystal structure of mouse RANKL-OPG complex | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 11, soluble form, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2012-03-12 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor.
Structure, 20, 2012
|
|
4K79
 
 | | Recognition of the Thomsen-Friedenreich Antigen by a Lamprey Variable Lymphocyte Receptor | | Descriptor: | Variable lymphocyte receptor, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Luo, M, Velikovsky, C.A, Yang, X.B, Mariuzza, R.A. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the thomsen-friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor.
J.Biol.Chem., 288, 2013
|
|
1NR7
 
 | | Crystal structure of apo bovine glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
3M19
 
 | |
1PK8
 
 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-06-05 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
2JV4
 
 | | Structure Characterisation of PINA WW Domain and Comparison with other Group IV WW Domains, PIN1 and ESS1 | | Descriptor: | Peptidyl-prolyl cis/trans isomerase | | Authors: | Ng, C.A, Kato, Y, Tanokura, M, Brownlee, R.T.C. | | Deposit date: | 2007-09-11 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural characterisation of PinA WW domain and a comparison with other Group IV WW domains, Pin1 and Ess1
Biochim.Biophys.Acta, 1784, 2008
|
|
4G26
 
 | | Crystal Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana with Ca | | Descriptor: | CALCIUM ION, Pentatricopeptide repeat-containing protein At2g32230, mitochondrial, ... | | Authors: | Koutmos, M, Howard, M.J, Fierke, C.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5' processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TYQ
 
 | |
2OZR
 
 | | MMP13 Catalytic Domain Complexed with 4-{[1-methyl-2,4-dioxo-6-(3-phenylprop-1-yn-1-yl)-1,4-dihydroquinazolin-3(2H)-yl]methyl}benzoic acid | | Descriptor: | 4-{[1-METHYL-2,4-DIOXO-6-(3-PHENYLPROP-1-YN-1-YL)-1,4-DIHYDROQUINAZOLIN-3(2H)-YL]METHYL}BENZOIC ACID, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Johnson, A.R, Pavlovsky, A.G, Ortwine, D.F, Prior, F, Man, C.-F, Bornemeier, D.A, Banotai, C.A, Mueller, W.T, McConnell, P, Yan, C.H, Baragi, V, Lesch, C, Roark, W.H, Lie, J.J, Fasquelle, V, Wilson, M, Robertson, D, Datta, K, Guzman, R, Han, H.-K, Dyer, R.D. | | Deposit date: | 2007-02-27 | | Release date: | 2007-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and characterization of a novel inhibitor of matrix metalloprotease-13 that reduces cartilage damage in vivo without joint fibroplasia side effects.
J.Biol.Chem., 282, 2007
|
|
4GIP
 
 | | Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 (PIV5) fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F1, Fusion glycoprotein F2 | | Authors: | Welch, B.D, Liu, Y, Kors, C.A, Leser, G.P, Jardetzky, T.S, Lamb, R.A. | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HPV
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-10-24 | | Release date: | 2012-11-14 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4HP9
 
 | |
3U4O
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannigrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4HIE
 
 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 | | Descriptor: | Antibody 023.102, Fab 023.102 | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
