4KJU
 
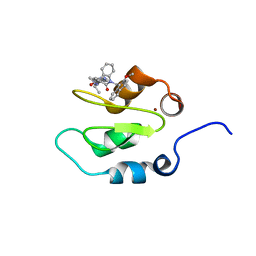 | | Crystal structure of XIAP-Bir2 with a bound benzodiazepinone inhibitor. | | Descriptor: | E3 ubiquitin-protein ligase XIAP, N-{(3S)-5-(4-aminobenzoyl)-1-[(2-methoxynaphthalen-1-yl)methyl]-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepin-3-yl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
6CS1
 
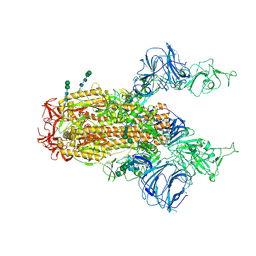 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
2M5K
 
 | | Atomic-resolution structure of a doublet cross-beta amyloid fibril | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.7 Å), SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-beta amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M5M
 
 | | Atomic-resolution structure of a triplet cross-beta amyloid fibril | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.2 Å), SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-beta amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
3SU4
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease,NS4A protein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SV8
 
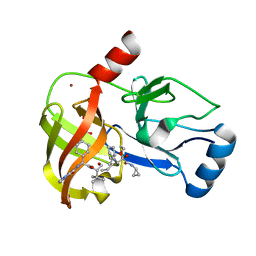 | | Crystal structure of NS3/4A protease variant D168A in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
2MZ9
 
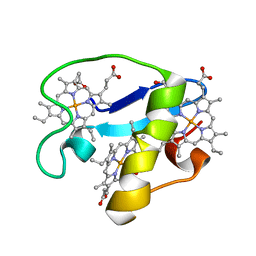 | | Solution structure of oxidized triheme cytochrome PpcA from Geobacter sulfurreducens | | Descriptor: | HEME C, PpcA | | Authors: | Morgado, L, Bruix, M, Pokkuluri, R, Salgueiro, C.A, Turner, D.L. | | Deposit date: | 2015-02-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Redox- and pH-linked conformational changes in triheme cytochrome PpcA from Geobacter sulfurreducens.
Biochem. J., 474, 2017
|
|
4L7Z
 
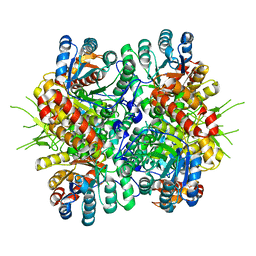 | | Crystal Structure of Chloroflexus aurantiacus malyl-CoA lyase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-14 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
4DQE
 
 | | Crystal Structure of (G16C/L38C) HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, Aspartyl protease | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
2FQX
 
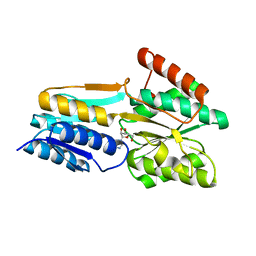 | | PnrA from Treponema pallidum complexed with guanosine | | Descriptor: | GUANOSINE, Membrane lipoprotein tmpC | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum
J.Biol.Chem., 281, 2006
|
|
4KX3
 
 | |
3SV6
 
 | | Crystal structure of NS3/4A protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
2N1L
 
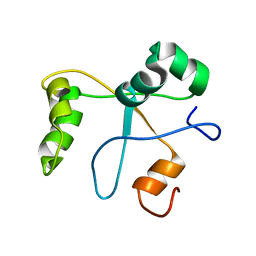 | | Solution structure of the BCOR PUFD | | Descriptor: | BCL-6 corepressor | | Authors: | Wong, S.J, Gearhart, M.D, Ha, D.J, Corcoran, C.M, Diaz, V, Taylor, A.B, Schirf, V, Ilangovan, U, Hinck, A.P, Demeler, B, Hart, J, Bardwell, V.J, Kim, C.A. | | Deposit date: | 2015-04-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the hierarchical assembly of the core of PRC1.1
To be Published
|
|
6CRX
 
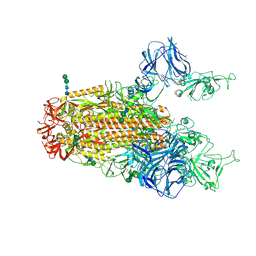 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CUL
 
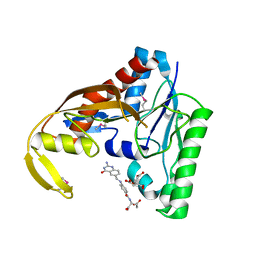 | | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase | | Descriptor: | CITRIC ACID, N-(4-{[(2-amino-4-oxo-1,4-dihydroquinazolin-6-yl)methyl]amino}benzene-1-carbonyl)-D-glutamic acid, Pyoverdine synthetase F | | Authors: | Kenjic, N, Hoag, M.R, Moraski, G.C, Caperelli, C.A, Moran, G.R, Lamb, A.L. | | Deposit date: | 2018-03-26 | | Release date: | 2019-02-06 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase.
Arch. Biochem. Biophys., 664, 2019
|
|
3LZ0
 
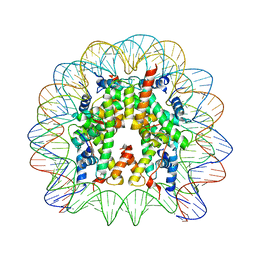 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 1) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
3LZ1
 
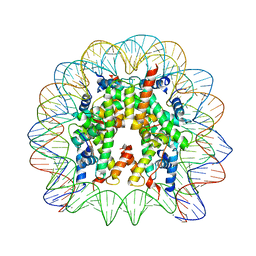 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 2) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
3LST
 
 | | Crystal Structure of CalO1, Methyltransferase in Calicheamicin Biosynthesis, SAH bound form | | Descriptor: | 1,2-ETHANEDIOL, CalO1 Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of CalO1: a putative orsellinic acid methyltransferase in the calicheamicin-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2G07
 
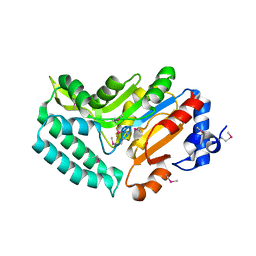 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, phospho-enzyme intermediate analog with Beryllium fluoride | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
6CRW
 
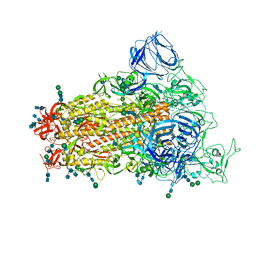 | | SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CSL
 
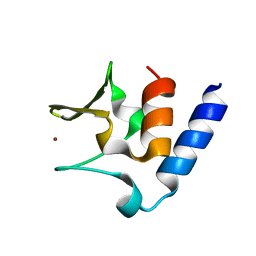 | | Pneumococcal PhtD protein 269-339 fragment with bound Zn(II) | | Descriptor: | Histidine triad protein D, ZINC ION | | Authors: | Luo, Z, Pederick, V.G, Paton, J.C, McDevitt, C.A, Kobe, B. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural characterisation of the HT3 motif of the polyhistidine triad protein D from Streptococcus pneumoniae.
FEBS Lett., 592, 2018
|
|
6VO3
 
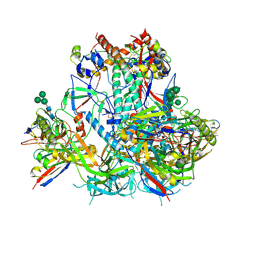 | | AMC009 SOSIP.v4.2 in complex with PGV04 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC009 SOSIP.v4.2 envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, de Val, N, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Neutralizing Antibody Responses Induced by HIV-1 Envelope Glycoprotein SOSIP Trimers Derived from Elite Neutralizers.
J.Virol., 94, 2020
|
|
4FN4
 
 | | short-chain NAD(H)-dependent dehydrogenase/reductase from Sulfolobus acidocaldarius | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Pennacchio, A, Sannino, V, Sorrentino, G, Rossi, M, Raia, C.A, Esposito, L. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural characterization of recombinant short-chain NAD(H)-dependent dehydrogenase/reductase from Sulfolobus acidocaldarius highly enantioselective on diaryl diketone benzil.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
2II4
 
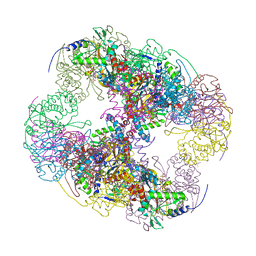 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Coenzyme A-bound form | | Descriptor: | CHLORIDE ION, COENZYME A, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
