5DIZ
 
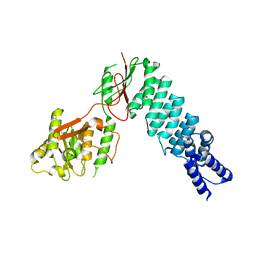 | | Crystal Structure of nuclear proteinaceous RNase P 2 (PRORP2) from A. thaliana | | Descriptor: | Proteinaceous RNase P 2, ZINC ION | | Authors: | Karasik, A, Shanmuganathan, A, Howard, M.J, Fierke, C.A, Koutmos, M. | | Deposit date: | 2015-09-01 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nuclear Protein-Only Ribonuclease P2 Structure and Biochemical Characterization Provide Insight into the Conserved Properties of tRNA 5' End Processing Enzymes.
J.Mol.Biol., 428, 2016
|
|
4WD1
 
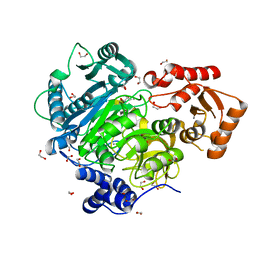 | | Acetoacetyl-CoA Synthetase from Streptomyces lividans | | Descriptor: | 1,2-ETHANEDIOL, Acetoacetate-CoA ligase, CALCIUM ION | | Authors: | Gulick, A.M, Mitchell, C.A. | | Deposit date: | 2014-09-05 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The structure of S. lividans acetoacetyl-CoA synthetase shows a novel interaction between the C-terminal extension and the N-terminal domain.
Proteins, 83, 2015
|
|
5K47
 
 | | CryoEM structure of the human Polycystin-2/PKD2 TRP channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Pike, A.C.W, Grieben, M, Shintre, C.A, Tessitore, A, Shrestha, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-20 | | Release date: | 2016-08-24 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2).
Nat. Struct. Mol. Biol., 24, 2017
|
|
4OD1
 
 | | Crystal structure of human Fab CAP256-VRC26.03, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.03 heavy chain, CAP256-VRC26.03 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4WB9
 
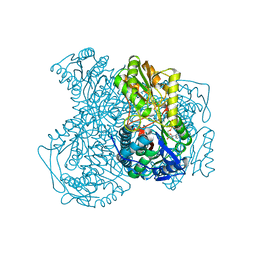 | | Human ALDH1A1 complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2014-09-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of a high-throughput in vitro assay to identify selective inhibitors for human ALDH1A1.
Chem.Biol.Interact., 234, 2015
|
|
5DIH
 
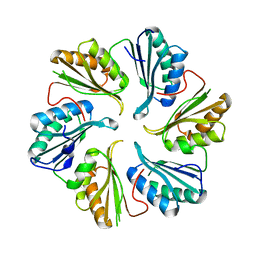 | |
5DII
 
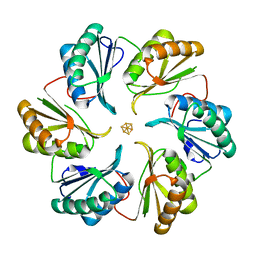 | | Structure of an engineered bacterial microcompartment shell protein binding a [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, Microcompartments protein | | Authors: | Sutter, M, Aussignargues, C, Turmo, A, Kerfeld, C.A. | | Deposit date: | 2015-09-01 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster.
J.Am.Chem.Soc., 138, 2016
|
|
4OX8
 
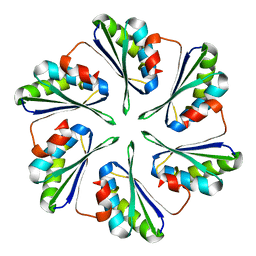 | |
4FFY
 
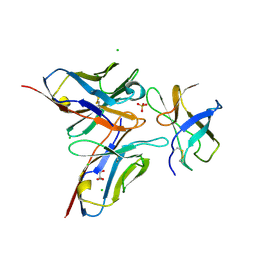 | | Crystal structure of DENV1-E111 single chain variable fragment bound to DENV-1 DIII, strain 16007. | | Descriptor: | CHLORIDE ION, DENV1-E111 single chain variable fragment (heavy chain), DENV1-E111 single chain variable fragment (light chain), ... | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
4FFZ
 
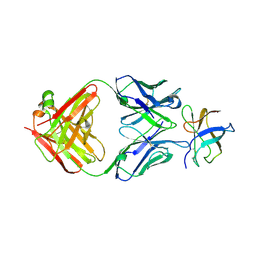 | | Crystal Structure of DENV1-E111 fab fragment bound to DENV-1 DIII (Western Pacific-74 strain). | | Descriptor: | DENV1-E111 fab fragment (heavy chain), DENV1-E111 fab fragment (light chain), Envelope protein E | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
3H5R
 
 | | Crystal structure of E. coli MccB + Succinimide | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H7C
 
 | | Crystal Structure of Arabidopsis thaliana Agmatine Deiminase from Cell Free Expression | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, Agmatine deiminase, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Arabidopsis thaliana Agmatine Deiminase
To be Published
|
|
3GNB
 
 | | Crystal structure of the RAG1 nonamer-binding domain with DNA | | Descriptor: | 5'-D(*AP*AP*TP*TP*TP*TP*CP*AP*GP*AP*AP*AP*CP*C)-3', 5'-D(*AP*GP*GP*TP*TP*TP*CP*TP*GP*AP*AP*AP*AP*C)-3', V(D)J recombination-activating protein 1 | | Authors: | Yin, F.F, Bailey, S, Innis, C.A, Steitz, T.A, Schatz, D.G. | | Deposit date: | 2009-03-16 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HAV
 
 | | Structure of the streptomycin-ATP-APH(2")-IIa ternary complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside phosphotransferase, MAGNESIUM ION, ... | | Authors: | Young, P.G, Baker, E.N, Vakulenko, S.B, Smith, C.A. | | Deposit date: | 2009-05-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2''-phosphotransferase-IIa [APH(2'')-IIa] provide insights into substrate selectivity in the APH(2'') subfamily.
J.Bacteriol., 191, 2009
|
|
7Z2Y
 
 | | Escherichia coli periplasmic phytase AppA T305E mutant, complex with myo-inositol hexakissulfate | | Descriptor: | Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, NICKEL (II) ION | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2W
 
 | | Escherichia coli periplasmic phytase AppA D304A,T305E mutant, complex with myo-inositol hexakissulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z1J
 
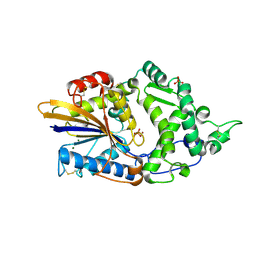 | | Escherichia coli periplasmic phytase AppA, complex with phosphate | | Descriptor: | Acidphosphatase, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2S
 
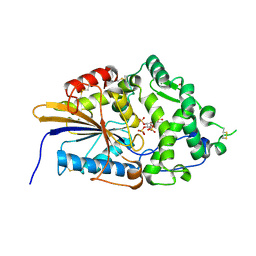 | | Escherichia coli periplasmic phytase AppA, complex with myo-inositol hexakissulfate | | Descriptor: | Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, NICKEL (II) ION, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z32
 
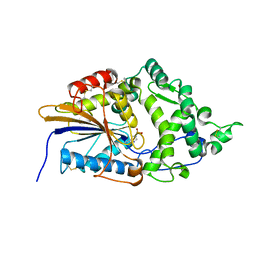 | | Escherichia coli periplasmic phytase AppA D304A mutant, phosphohistidine intermediate | | Descriptor: | Acidphosphatase, NICKEL (II) ION | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2T
 
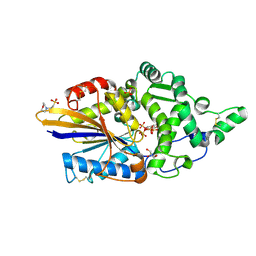 | | Escherichia coli periplasmic phytase AppA D304A mutant, complex with myo-inositol hexakissulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
5DYR
 
 | | Structure of virulence-associated protein D (VapD) from Xylella fastidiosa | | Descriptor: | Virulence-associated protein D | | Authors: | Kochneva, M.V, dos Santos, M.L, dos Santos, C.A, de Souza, A.P, Polikarpov, I, Aparicio, R, Golubev, A.M. | | Deposit date: | 2015-09-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of virulence-associated protein D (VapD) from Xylella fastidiosa
To Be Published
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4FFE
 
 | | The structure of cowpox virus CPXV018 (OMCP) | | Descriptor: | CPXV018 protein | | Authors: | Lazear, E, Peterson, L.W, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Cowpox Virus-Encoded NKG2D Ligand OMCP.
J.Virol., 87, 2013
|
|
4FFB
 
 | | A TOG:alpha/beta-tubulin Complex Structure Reveals Conformation-Based Mechanisms For a Microtubule Polymerase | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Ye, X, Huddleston, P, Brautigam, C.A, Rice, L.M. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | A TOG: alpha beta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase.
Science, 337, 2012
|
|
4FG0
 
 | |
