1SQX
 
 | | Crystal Structure Analysis of Bovine Bc1 with Stigmatellin A | | Descriptor: | Cytochrome b, Cytochrome c1, heme protein, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-21 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
3BVO
 
 | | Crystal structure of human co-chaperone protein HscB | | Descriptor: | Co-chaperone protein HscB, mitochondrial precursor, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-01-07 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain.
J.Biol.Chem., 283, 2008
|
|
3BOM
 
 | | Crystal structure of trout hemoglobin at 1.35 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Hemoglobin subunit alpha-4, Hemoglobin subunit beta-4, ... | | Authors: | Aranda IV, R, Bingman, C.A, Bitto, E, Wesenberg, G.E, Richards, M, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Trout Hemoglobin Crystal Structure at 1.35 Angstroms Resolution.
To be Published
|
|
5TSS
 
 | | TOXIC SHOCK SYNDROME TOXIN-1: ORTHORHOMBIC P222(1) CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
3BUJ
 
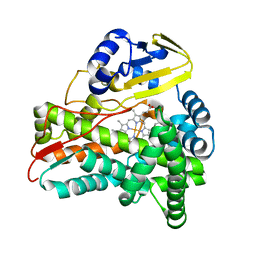 | | Crystal Structure of CalO2 | | Descriptor: | CalO2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McCoy, J.G, Johnson, H.D, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway.
Proteins, 74, 2009
|
|
1J4V
 
 | | CYANOVIRIN-N | | Descriptor: | CYANOVIRIN-N | | Authors: | Clore, G.M, Bewley, C.A. | | Deposit date: | 2001-11-21 | | Release date: | 2002-03-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Using conjoined rigid body/torsion angle simulated annealing to determine the relative orientation of covalently linked protein domains from dipolar couplings.
J.Magn.Reson., 154, 2002
|
|
2SEC
 
 | |
8OXE
 
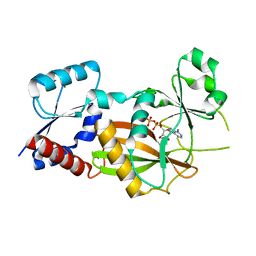 | | Inositol 1,3,4-trisphosphate 5/6-kinase 1 from Solanum tuberosum (StITPK1) in complex with ADP/Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-tetrakisphosphate 1-kinase, MAGNESIUM ION | | Authors: | Faba-Rodriguez, R, Li, A.W.H, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2023-05-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure and Enzymology of Solanum tuberosum Inositol Tris/Tetrakisphosphate Kinase 1 ( St ITPK1).
Biochemistry, 63, 2024
|
|
8PNL
 
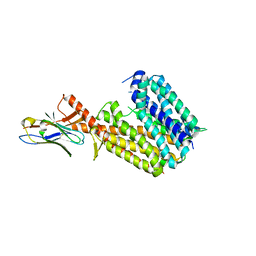 | | Outward-open conformation of a Major Facilitator Superfamily (MFS) transporter MHAS2168, a homologue of Rv1410 from M. tuberculosis, in complex with an alpaca nanobody | | Descriptor: | Nb_H2, Putative triacylglyceride transporter | | Authors: | Remm, S, Schoeppe, J, Hutter, C.A.J, Gonda, I, Seeger, M.A. | | Deposit date: | 2023-06-30 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for triacylglyceride extraction from mycobacterial inner membrane by MFS transporter Rv1410.
Nat Commun, 14, 2023
|
|
8PAZ
 
 | | OXIDIZED NATIVE PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-24 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
1IAO
 
 | | CLASS II MHC I-AD IN COMPLEX WITH OVALBUMIN PEPTIDE 323-339 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-AD | | Authors: | Scott, C.A, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 1998-03-13 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues.
Immunity, 8, 1998
|
|
8P94
 
 | |
8QKT
 
 | |
1LCF
 
 | | CRYSTAL STRUCTURE OF COPPER-AND OXALATE-SUBSTITUTED HUMAN LACTOFERRIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, COPPER (II) ION, ... | | Authors: | Smith, C.A, Anderson, B.F, Baker, H.M, Baker, E.N. | | Deposit date: | 1994-01-11 | | Release date: | 1994-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of copper- and oxalate-substituted human lactoferrin at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1SQP
 
 | | Crystal Structure Analysis of Bovine Bc1 with Myxothiazol | | Descriptor: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-19 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
1SQQ
 
 | | Crystal Structure Analysis of Bovine Bc1 with Methoxy Acrylate Stilbene (MOAS) | | Descriptor: | Cytochrome b, Cytochrome c1, heme protein, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-19 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
8GFT
 
 | | Hsp90 provides platform for CRaf dephosphorylation by PP5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, ... | | Authors: | Jaime-Garza, M, Nowotny, C.A, Coutandin, D, Wang, F, Tabios, M, Agard, D.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Hsp90 provides a platform for kinase dephosphorylation by PP5.
Nat Commun, 14, 2023
|
|
6E3Y
 
 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
6EDM
 
 | | Structure of apo-CDD-1 beta-lactamase | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-14 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of CDD-1, the intrinsic class D beta-lactamase from the pathogenic Gram-positive bacterium Clostridioides difficile, and its complex with cefotaxime.
J.Struct.Biol., 208, 2019
|
|
6EFU
 
 | | Crystal structure of the double mutant L167W / P172L of the beta-glucosidase from Trichoderma harzianum | | Descriptor: | Beta-glucosidase, NITRATE ION | | Authors: | Morais, M.A.B, Santos, C.A, Tonoli, C.C.C, Souza, A.P, Murakami, M.T. | | Deposit date: | 2018-08-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An engineered GH1 beta-glucosidase displays enhanced glucose tolerance and increased sugar release from lignocellulosic materials.
Sci Rep, 9, 2019
|
|
4LRC
 
 | | Phosphopentomutase V158L variant | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
1HNK
 
 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III, APO TETRAGONAL FORM | | Descriptor: | BETA-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Head, M, Lonsdale, J, Konstantinidis, A.K. | | Deposit date: | 2000-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of beta-ketoacyl-acyl carrier protein synthase III.
J.Mol.Biol., 307, 2001
|
|
4M8N
 
 | | Crystal Structure of PlexinC1/Rap1B Complex | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pascoe, H.G, Wang, Y, Brautigam, C.A, He, H, Zhang, X. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin.
Elife, 2, 2013
|
|
6E47
 
 | |
6E48
 
 | | Crystal Structure of the Murine Norovirus VP1 P domain in complex with the CD300lf Receptor and Lithocholic Acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
