7N22
 
 | | NMR structure of AnIB[Y(SO3)16Y]-NH2 | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N45
 
 | |
7N0T
 
 | | NMR structure of EpI[Y(SO)315Y]-OH | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N23
 
 | | NMR structure of AnIB[Y(SO3)16Y]-OH | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
5Z1Y
 
 | | mBjAMP1 structure | | Descriptor: | mBjAMP1 peptide | | Authors: | Nam, J.Y, Lee, C.W. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | mBjAMP1 structure
To Be Published
|
|
7N20
 
 | | NMR structure of native AnIB | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
5Z2O
 
 | |
7NQ7
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH (S)-N-ethyl-3-(1-methyl-1H-1,2,3-triazol-4-yl)-4-(1-phenylethoxy)benzamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ~{N}-ethyl-3-(1-methyl-1,2,3-triazol-4-yl)-4-[(1~{S})-1-phenylethoxy]benzamide | | Authors: | Chung, C. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NSE
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, ADMA COMPLEX | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2002-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of the Heme Domain of Bovine Endothelial Nitric Oxide Synthase Complexed with Arginine Analogues
To be Published
|
|
7NQ5
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 3-(2-(benzyloxy)phenyl)-5-methyl-1H-1,2,4-triazole | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-methyl-5-(2-phenylmethoxyphenyl)-4~{H}-1,2,4-triazole, ... | | Authors: | Chung, C. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NQ8
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-ethyl-3-(1-methyl-1H-1,2,3-triazol-4-yl)-4-(pyridin-2-ylmethoxy)benzamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-ethyl-3-(1-methyl-1H-1,2,3-triazol-4-yl)-4-(pyridin-2-ylmethoxy)benzamide | | Authors: | Chung, C.W. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NQI
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 2-benzyl-N-cyclopropyl-6-(1-methyl-1H-1,2,3-triazol-4-yl)isonicotinamide | | Descriptor: | 1,2-ETHANEDIOL, 2-benzyl-N-cyclopropyl-6-(1-methyl-1H-1,2,3-triazol-4-yl)isonicotinamide, Bromodomain-containing protein 2 | | Authors: | Chung, C.W. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NQ9
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 2-benzyl-N-cyclopropyl-6-(1-methyl-1H-1,2,3-triazol-4-yl)isonicotinamide | | Descriptor: | 1,2-ETHANEDIOL, 6-benzyl-N4-((1r,3r)-3-hydroxycyclobutyl)-N2-methylpyridine-2,4-dicarboxamide, Bromodomain-containing protein 2 | | Authors: | Chung, C.W. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B5D
 
 | |
7NQJ
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-ethyl-2-(1-methyl-1H-1,2,3-triazol-4-yl)-6-(1-phenylethyl)isonicotinamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chung, C. | | Deposit date: | 2021-03-01 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NRN
 
 | | NMR structure of GIPC1-GH2 domain | | Descriptor: | PDZ domain-containing protein GIPC1 | | Authors: | Barthe, P, Roumestand, C. | | Deposit date: | 2021-03-04 | | Release date: | 2021-04-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pressure and Chemical Unfolding of an alpha-Helical Bundle Protein: The GH2 Domain of the Protein Adaptor GIPC1.
Int J Mol Sci, 22, 2021
|
|
7B06
 
 | | TgoT_RT521 apo | | Descriptor: | DNA polymerase | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
5ZO5
 
 | | active state of the nuclease | | Descriptor: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
7NRI
 
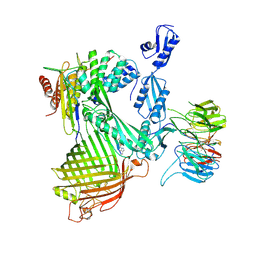 | | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
6Q1C
 
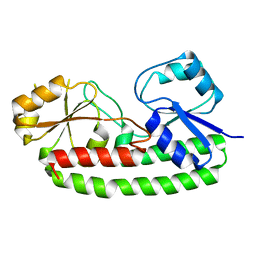 | |
5ZLJ
 
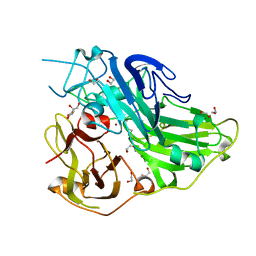 | | Crystal structure of CotA native enzyme, PH8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
5ZHQ
 
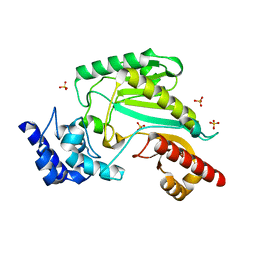 | |
7NPR
 
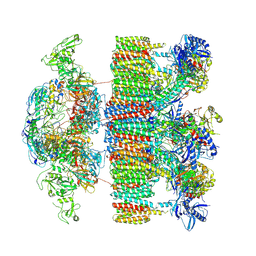 | | Structure of an intact ESX-5 inner membrane complex, Composite C3 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
6Q1J
 
 | |
8HE4
 
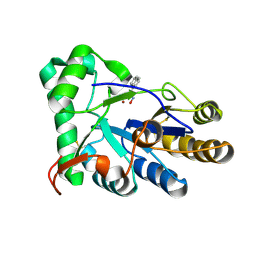 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
