2AP9
 
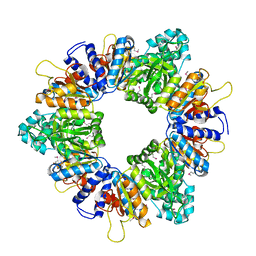 | | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551 | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, acetylglutamate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-15 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551
To be Published
|
|
2AWD
 
 | |
2B5G
 
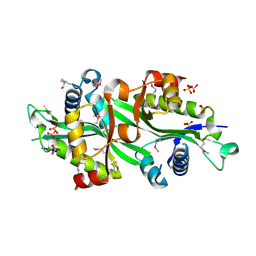 | | Wild Type SSAT- 1.7A structure | | Descriptor: | Diamine acetyltransferase 1, SULFATE ION | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2F1R
 
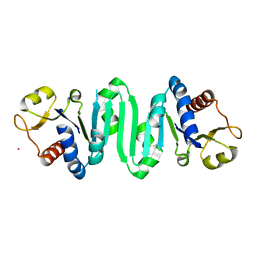 | | Crystal Structure of molybdopterin-guanine biosynthesis protein B (mobB) | | Descriptor: | CHLORIDE ION, PRASEODYMIUM ION, molybdopterin-guanine dinucleotide biosynthesis protein B (mobB) | | Authors: | Damodharan, L, Eswaramoorthy, S, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of molybdopterin-guanine dinucleotide biosynthesis protein B (mobB)
To be Published
|
|
2EUI
 
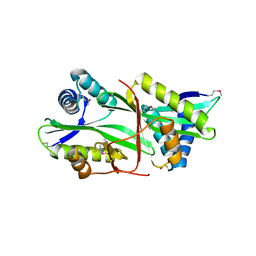 | |
2ESR
 
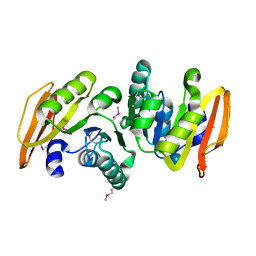 | | conserved hypothetical protein- streptococcus pyogenes | | Descriptor: | Methyltransferase, alpha-D-glucopyranose | | Authors: | Jiang, J, Min, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-10-26 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of hypothetical protein of Streptococcus Pygenes
To be Published
|
|
2F02
 
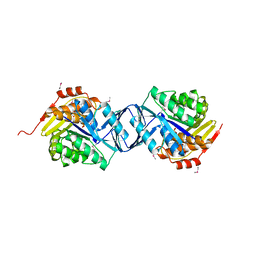 | |
2FIQ
 
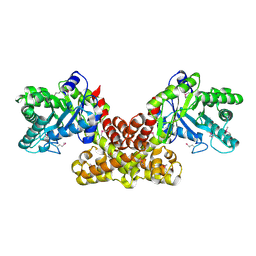 | |
2F7F
 
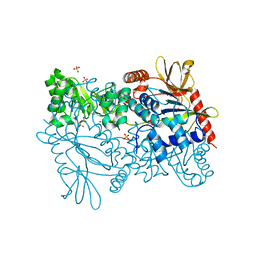 | | Crystal structure of Enterococcus faecalis putative nicotinate phosphoribosyltransferase, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | | Descriptor: | DIPHOSPHATE, GLYCEROL, NICOTINIC ACID, ... | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Enterococcus Faecalis Nicotinate Phosphoribosyltransferase
To be Published
|
|
2FGS
 
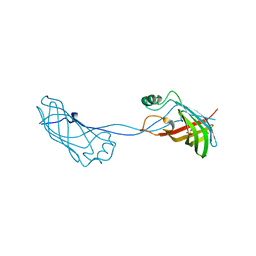 | | Crystal structure of Campylobacter jejuni YCEI protein, structural genomics | | Descriptor: | Putative periplasmic protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Campylobacter Jejuni YceI Periplasmic Protein
To be Published
|
|
1CI0
 
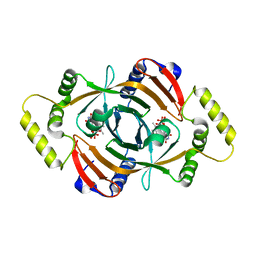 | | PNP OXIDASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (PNP OXIDASE) | | Authors: | Shi, W, Ostrov, D.A, Gerchman, S.E, Graziano, V, Kycia, H, Studier, B, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-04-06 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of PNP Oxidase from S. Cerevisiae
To be Published
|
|
1F89
 
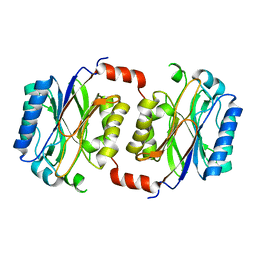 | | Crystal structure of Saccharomyces cerevisiae Nit3, a member of branch 10 of the nitrilase superfamily | | Descriptor: | 32.5 KDA PROTEIN YLR351C | | Authors: | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-06-29 | | Release date: | 2001-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative CN hydrolase from yeast
Proteins, 52, 2003
|
|
1DFC
 
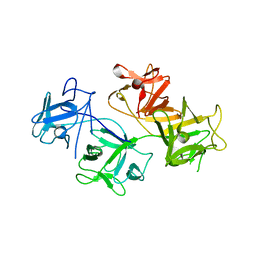 | | CRYSTAL STRUCTURE OF HUMAN FASCIN, AN ACTIN-CROSSLINKING PROTEIN | | Descriptor: | FASCIN | | Authors: | Fedorov, A.A, Fedorov, E.V, Ono, S, Matsumura, F, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-11-18 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein.
J.Mol.Biol., 400, 2010
|
|
1CT5
 
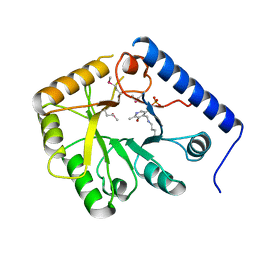 | | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YBL036C-SELENOMET CRYSTAL | | Descriptor: | PROTEIN (YEAST HYPOTHETICAL PROTEIN, SELENOMET), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3IK4
 
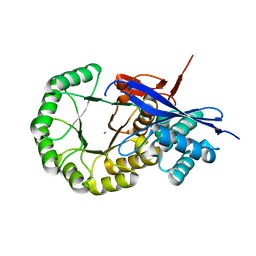 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Herpetosiphon aurantiacus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing protein, POTASSIUM ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Iizuka, M, Sauder, J.M, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5Q0M
 
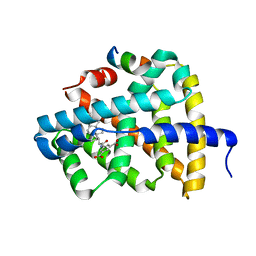 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-{[(3beta,5beta,14beta,17alpha)-3-hydroxy-24-oxocholan-24-yl]amino}benzene-1,3-dicarboxylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0W
 
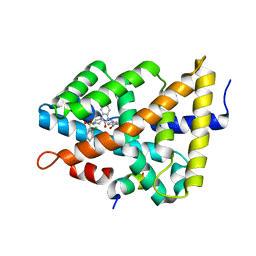 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-({5-bromo-1'-[(2-chlorophenyl)sulfonyl]-2-oxospiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, cDNA FLJ76652, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q12
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-(2,6-difluorophenyl)-N-(2,6-dimethylphenyl)-5-methylimidazo[1,2-a]pyridin-3-amine, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1G
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2E)-N-cyclohexyl-N-(cyclohexylcarbamoyl)-3-(4-fluorophenyl)prop-2-enamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0O
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-{2-[4-(benzenecarbonyl)phenyl]-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, CHLORIDE ION, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q14
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}-3-fluorobenzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1E
 
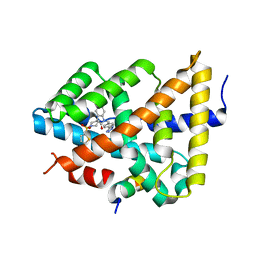 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-bromo-1-{[4-(1H-tetrazol-5-yl)phenyl]methyl}-1'-(thiophene-2-sulfonyl)spiro[indole-3,4'-piperidin]-2(1H)-one, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0U
 
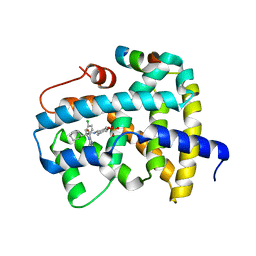 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)cyclohexyl hydrogen sulfate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1C
 
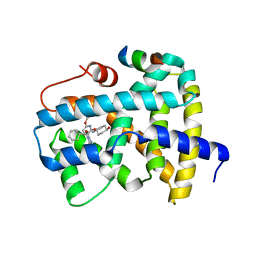 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0N
 
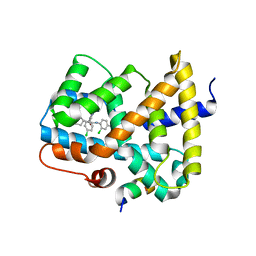 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 3-chloro-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
