1G7S
 
 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1G61
 
 | | CRYSTAL STRUCTURE OF M.JANNASCHII EIF6 | | Descriptor: | TRANSLATION INITIATION FACTOR 6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
1G7R
 
 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B | | Descriptor: | TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1EJ4
 
 | | COCRYSTAL STRUCTURE OF EIF4E/4E-BP1 PEPTIDE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E BINDING PROTEIN 1 | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-02-28 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G.
Mol.Cell, 3, 1999
|
|
1G7T
 
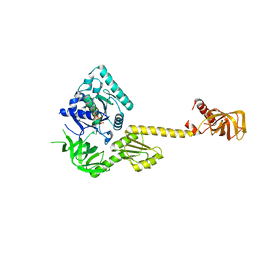 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1G62
 
 | | CRYSTAL STRUCTURE OF S.CEREVISIAE EIF6 | | Descriptor: | RIBOSOME ANTI-ASSOCIATION FACTOR EIF6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
1DJ8
 
 | |
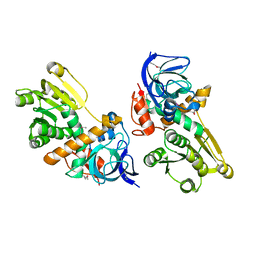
3GAZ
 
 | |
3GBT
 
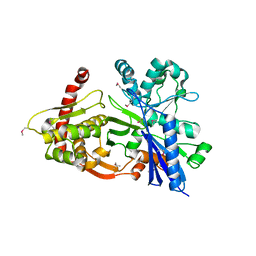 | |
1SG9
 
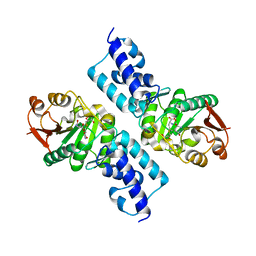 | | Crystal structure of Thermotoga maritima protein HEMK, an N5-glutamine methyltransferase | | Descriptor: | GLUTAMINE, S-ADENOSYLMETHIONINE, hemK protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel mode of dimerization via formation of a glutamate anhydride crosslink in a protein crystal structure.
Proteins, 71, 2008
|
|
3GG2
 
 | | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate | | Descriptor: | Sugar dehydrogenase, UDP-glucose/GDP-mannose dehydrogenase family, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate
To be Published
|
|
3GBW
 
 | | Crystal structure of the first PHR domain of the Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Klemke, R.L, Miller, S.A, Bain, K.T, Rutter, M.E, Tarun, G, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
3GH1
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | PHOSPHATE ION, Predicted nucleotide-binding protein | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae.
To be Published
|
|
1SGM
 
 | |
3GV1
 
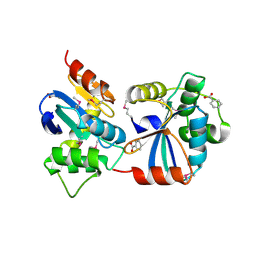 | |
3H0U
 
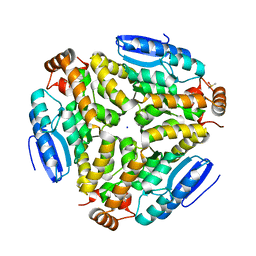 | | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis | | Descriptor: | DIMETHYL SULFOXIDE, Putative enoyl-CoA hydratase, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis
To be Published
|
|
3GL3
 
 | |
3GV0
 
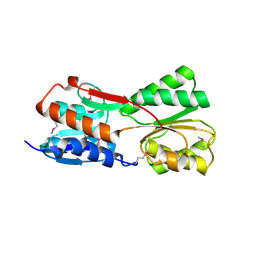 | |
3GY1
 
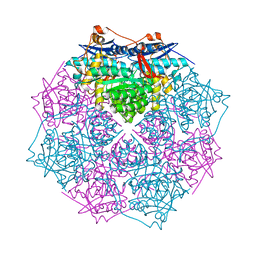 | | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium beijerinckii NCIMB 8052 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium
beijerinckii NCIMB 8052
To be Published
|
|
3H5T
 
 | |
3GPV
 
 | | Crystal structure of a transcriptional regulator, MerR family from Bacillus thuringiensis | | Descriptor: | Transcriptional regulator, MerR family | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-23 | | Release date: | 2009-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a transcriptional regulator, MerR family from Bacillus thuringiensis
To be Published
|
|
3GKB
 
 | | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis
To be Published
|
|
3GNL
 
 | | Structure of uncharacterized protein (LMOf2365_1472) from Listeria monocytogenes serotype 4b | | Descriptor: | uncharacterized protein, DUF633, LMOf2365_1472 | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-17 | | Release date: | 2009-04-21 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of uncharacterized protein (LMOf2365_1472) from
Listeria monocytogenes serotype 4b
To be Published
|
|
3GP4
 
 | | Crystal structure of putative MerR family transcriptional regulator from listeria monocytogenes | | Descriptor: | D-METHIONINE, GLYCEROL, SODIUM ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of putative MerR family transcriptional regulator from Listeria monocytogenes
To be published
|
|
3GT7
 
 | | CRYSTAL STRUCTURE OF SIGNAL RECEIVER DOMAIN OF SIGNAL TRANSDUCTION HISTIDINE KINASE FROM Syntrophus aciditrophicus | | Descriptor: | Sensor protein | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Hu, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Signal Transduction Kinase from Syntrophus Aciditrophicus
To be Published
|
|
