4IJI
 
 | | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione | | Descriptor: | ACRYLIC ACID, BENZOIC ACID, Glutathione S-transferase-like protein YibF, ... | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione
To be Published
|
|
4JHM
 
 | | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Pseudovibrio sp. | | Descriptor: | Mandelate racemase / muconate lactonizing enzyme, C-terminal domain protein | | Authors: | Hegde, R.P, Toro, R, Burley, S.K, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Pseudovibrio sp.
To be published
|
|
4ID0
 
 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione sulfinic acid (gso2h) and acetate | | Descriptor: | ACETATE ION, GLYCEROL, Glutathione S-transferase-like protein YibF, ... | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-11 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione sulfinic acid (gso2h) and acetate
To be Published
|
|
4IBP
 
 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase-like protein YibF, SULFATE ION | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target IFI-900011, with bound glutathione
To be Published
|
|
3SMD
 
 | |
3SN4
 
 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate
To be Published
|
|
3T8L
 
 | | Crystal Structure of adenine deaminase with Mn/Fe | | Descriptor: | Adenine deaminase 2, UNKNOWN ATOM OR ION | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-08-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The catalase activity of diiron adenine deaminase.
Protein Sci., 20, 2011
|
|
3T81
 
 | |
3TVI
 
 | | Crystal structure of Clostridium acetobutylicum aspartate kinase (CaAK): An important allosteric enzyme for industrial amino acids production | | Descriptor: | ASPARTIC ACID, Aspartokinase, LYSINE | | Authors: | Manjasetty, B.A, Chance, M.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-09-20 | | Release date: | 2011-11-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Clostridium acetobutylicum Aspartate kinase (CaAK): An important allosteric enzyme for amino acids production.
Biotechnol Rep (Amst), 3, 2014
|
|
1I2T
 
 | |
1I9A
 
 | | STRUCTURAL STUDIES OF CHOLESTEROL BIOSYNTHESIS: MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE AND ISOPENTENYL DIPHOSPHATE ISOMERASE | | Descriptor: | ISOPENTENYL-DIPHOSPHATE DELTA-ISOMERASE, MANGANESE (II) ION | | Authors: | Bonanno, J.B, Edo, C, Eswar, N, Pieper, U, Romanowski, M.J, Ilyin, V, Gerchman, S.E, Kycia, H, Studier, F.W, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-03-18 | | Release date: | 2001-03-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1AM9
 
 | | HUMAN SREBP-1A BOUND TO LDL RECEPTOR PROMOTER | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*AP*TP*CP*AP*CP*CP*CP*CP*AP*CP*T P*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*GP*TP*GP*GP*GP*GP*TP*GP*AP*TP*CP*T )-3'), MAGNESIUM ION, ... | | Authors: | Parraga, A, Burley, S.K. | | Deposit date: | 1997-06-25 | | Release date: | 1998-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution.
Structure, 6, 1998
|
|
1JFI
 
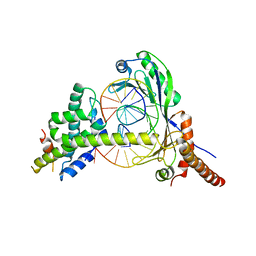 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | Descriptor: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | Authors: | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-20 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
1BH0
 
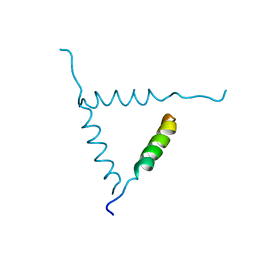 | | STRUCTURE OF A GLUCAGON ANALOG | | Descriptor: | GLUCAGON | | Authors: | Sturm, N.S, Lin, Y, Burley, S.K, Krstenansky, J.L, Ahn, J.-M, Azizeh, B.Y, Trivedi, D, Hruby, V.J. | | Deposit date: | 1998-06-11 | | Release date: | 1998-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies on positions 17, 18, and 21 replacement analogues of glucagon: the importance of charged residues and salt bridges in glucagon biological activity.
J.Med.Chem., 41, 1998
|
|
1JMT
 
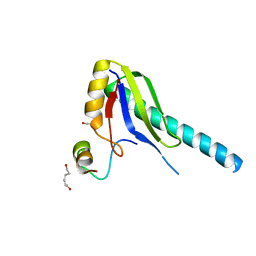 | | X-ray Structure of a Core U2AF65/U2AF35 Heterodimer | | Descriptor: | HEXANE-1,6-DIOL, SPLICING FACTOR U2AF 35 KDA SUBUNIT, SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Kielkopf, C.L, Rodionova, N.A, Green, M.R, Burley, S.K. | | Deposit date: | 2001-07-19 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JSS
 
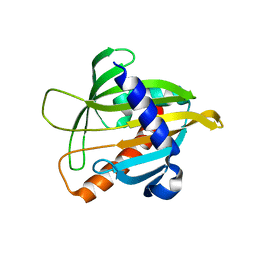 | | Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4). | | Descriptor: | cholesterol-regulated START protein 4 | | Authors: | Romanowski, M.J, Soccio, R.E, Breslow, J.L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-08-17 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JSX
 
 | |
1JYH
 
 | |
1JR7
 
 | |
1BO4
 
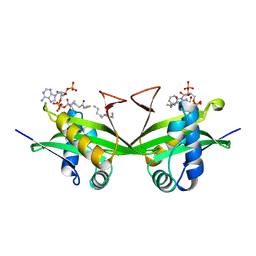 | | CRYSTAL STRUCTURE OF A GCN5-RELATED N-ACETYLTRANSFERASE: SERRATIA MARESCENS AMINOGLYCOSIDE 3-N-ACETYLTRANSFERASE | | Descriptor: | COENZYME A, PROTEIN (SERRATIA MARCESCENS AMINOGLYCOSIDE-3-N-ACETYLTRANSFERASE), SPERMIDINE | | Authors: | Wolf, E, Vassilev, A, Makino, Y, Sali, A, Nakatani, Y, Burley, S.K. | | Deposit date: | 1998-08-08 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase.
Cell(Cambridge,Mass.), 94, 1998
|
|
1JG8
 
 | | Crystal Structure of Threonine Aldolase (Low-specificity) | | Descriptor: | CALCIUM ION, L-allo-threonine aldolase, SODIUM ION | | Authors: | Kielkopf, C.L, Bonanno, J, Ray, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-23 | | Release date: | 2001-07-04 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Low-specificity Threonine Aldolase, a Key Enzyme in Glycine Biosynthesis
To be published
|
|
1JD1
 
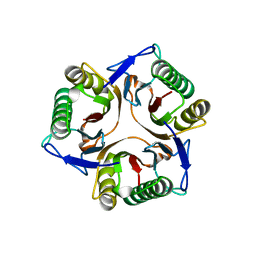 | |
1KNZ
 
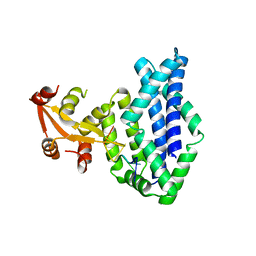 | | Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer | | Descriptor: | 5'-R(*UP*GP*AP*CP*C)-3', Nonstructural RNA-binding Protein 34 | | Authors: | Deo, R.C, Groft, C.M, Rajashankar, K.R, Burley, S.K. | | Deposit date: | 2001-12-19 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer.
Cell(Cambridge,Mass.), 108, 2002
|
|
1KUY
 
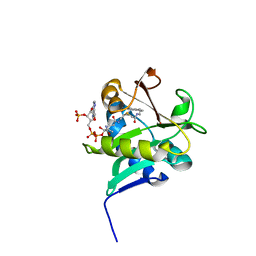 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
1KCX
 
 | | X-ray structure of NYSGRC target T-45 | | Descriptor: | DIHYDROPYRIMIDINASE RELATED PROTEIN-1 | | Authors: | Deo, R.C, Schmidt, E.F, Strittmatter, S.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-11-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for CRMP function in plexin-dependent semaphorin3A signaling
Embo J., 23, 2004
|
|
