4OQW
 
 | | Crystal structure of mCardinal far-red fluorescent protein | | Descriptor: | Fluorescent protein FP480 | | Authors: | Burg, J.S, Chu, J, Lam, A.J, Lin, M.Z, Garcia, K.C. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-12 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein.
Nat.Methods, 11, 2014
|
|
4XT3
 
 | | Structure of a viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fractalkine, G-protein coupled receptor homolog US28, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
4XT1
 
 | | Structure of a nanobody-bound viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Fractalkine, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
8AFF
 
 | |
8AFG
 
 | |
5WB1
 
 | |
5WB2
 
 | | US28 bound to engineered chemokine CX3CL1.35 and nanobodies | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Jude, K.M, Burg, J.S, Tsutsumi, N, Miles, T.F, Garcia, K.C. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Viral GPCR US28 can signal in response to chemokine agonists of nearly unlimited structural degeneracy.
Elife, 7, 2018
|
|
4UJD
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B PRE-like state | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Eif5B on Hcv Ires
Nat.Struct.Mol.Biol., 21, 2014
|
|
4UJC
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B POST-like state | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Initiation Factor 5B on Hcv-Ires RNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
8ATD
 
 | | Wild type hexamer oxalyl-CoA synthetase (OCS) | | Descriptor: | Oxalate--CoA ligase | | Authors: | Lill, P, Burgi, J, Raunser, S, Wilmanns, M, Gatsogiannis, C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol.Chem., 404, 2023
|
|
6Y5S
 
 | | Crystal structure of savinase at cryogenic conditions | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
6Y5T
 
 | | Crystal structure of savinase at room temperature | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
6HHC
 
 | | Allosteric Inhibition as a new mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated form of Coagulation Factor XI | | Descriptor: | Coagulation factor XI, DIMETHYL SULFOXIDE, FXIA ANTIBODY FAB HEAVY CHAIN, ... | | Authors: | Schaefer, M, Buchmueller, A, Dittmer, F, Strassburger, J, Wilmen, A. | | Deposit date: | 2018-08-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric Inhibition as a New Mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated Form of Coagulation Factor XI.
J.Mol.Biol., 431, 2019
|
|
5FLX
 
 | | Mammalian 40S HCV-IRES complex | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yamamoto, H, Collier, M, Loerke, J, Ismer, J, Schmidt, A, Hilal, T, Sprink, T, Yamamoto, K, Mielke, T, Burger, J, Shaikh, T.R, Dabrowski, M, Hildebrand, P.W, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Architecture of the Ribosome-Bound Hepatitis C Virus Internal Ribosomal Entry Site RNA.
Embo J., 34, 2015
|
|
6H75
 
 | | SiaP A11N in complex with Neu5Ac (RT) | | Descriptor: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein SiaP | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6H76
 
 | | SiaP in complex with Neu5Ac (RT) | | Descriptor: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
5AJ0
 
 | | Cryo electron microscopy of actively translating human polysomes (POST state). | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Behrmann, E, Loerke, J, Budkevich, T.V, Yamamoto, K, Schmidt, A, Penczek, P.A, Vos, M.R, Burger, J, Mielke, T, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Snapshots of Actively Translating Human Ribosomes
Cell(Cambridge,Mass.), 161, 2015
|
|
5DYN
 
 | | B. fragilis cysteine protease | | Descriptor: | CHLORIDE ION, Putative peptidase, SODIUM ION | | Authors: | Choi, V.M, Herrou, J, Hecht, A.L, Turner, J.R, Crosson, S, Bubeck Wardenburg, J. | | Deposit date: | 2015-09-24 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice.
Nat. Med., 22, 2016
|
|
3R15
 
 | | Structure Treponema Denticola Factor H Binding Protein | | Descriptor: | Factor H binding protein, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Burgner, J, Heroux, A, Bell, J.K, Marconi, R.T. | | Deposit date: | 2011-03-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure Treponema Denticola Factor H Binding Protein
To be Published
|
|
3K30
 
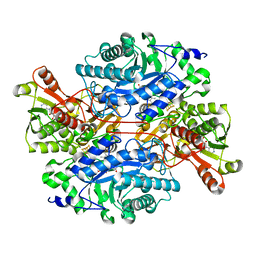 | | Histamine dehydrogenase from Nocardiodes simplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Histamine dehydrogenase, ... | | Authors: | Scott, E.E, Reed, T.M, Limburg, J. | | Deposit date: | 2009-09-30 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of histamine dehydrogenase from Nocardioides simplex.
J.Biol.Chem., 285, 2010
|
|
3ITQ
 
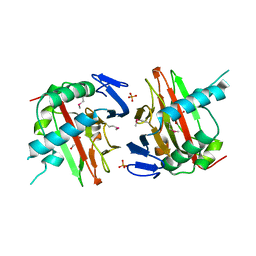 | | Crystal Structure of a Prolyl 4-Hydroxylase from Bacillus anthracis | | Descriptor: | GLYCEROL, PHOSPHATE ION, Prolyl 4-hydroxylase, ... | | Authors: | Culpepper, M.A, Scott, E.E, Limburg, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-12-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of prolyl 4-hydroxylase from Bacillus anthracis.
Biochemistry, 49, 2010
|
|
2CJW
 
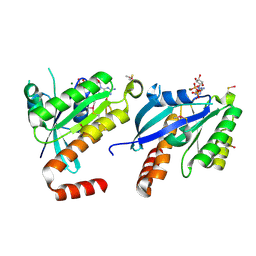 | | Crystal structure of the small GTPase Gem (GemDNDCaM) in complex to Mg.GDP | | Descriptor: | GTP-BINDING PROTEIN GEM, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Splingard, A, Menetrey, J, Perderiset, M, Cicolari, J, Hamoudi, F, Cabanie, L, El Marjou, A, Wells, A, Houdusse, A, de Gunzburg, J. | | Deposit date: | 2006-04-09 | | Release date: | 2006-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of the Gem Gtpase.
J.Biol.Chem., 282, 2007
|
|
5LWH
 
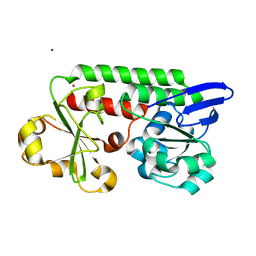 | | CeuE (Y288F variant) a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin ABC transporter substrate-binding protein, ZINC ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
1RPU
 
 | | Crystal Structure of CIRV p19 bound to siRNA | | Descriptor: | 19 kDa protein, 5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*UP*CP*AP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3' | | Authors: | Vargason, J.M, Szittya, G, Burgyan, J, Hall, T.M.T. | | Deposit date: | 2003-12-03 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Size selective recognition of siRNA by an RNA silencing suppressor
Cell(Cambridge,Mass.), 115, 2003
|
|
