6RQF
 
 | | 3.6 Angstrom cryo-EM structure of the dimeric cytochrome b6f complex from Spinacia oleracea with natively bound thylakoid lipids and plastoquinone molecules | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Malone, L.A, Qian, P, Mayneord, G.E, Hitchcock, A, Farmer, D, Thompson, R, Swainsbury, D.J.K, Ranson, N, Hunter, C.N, Johnson, M.P. | | Deposit date: | 2019-05-15 | | Release date: | 2019-11-13 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structure of the spinach cytochrome b6f complex at 3.6 angstrom resolution.
Nature, 575, 2019
|
|
1TKF
 
 | | Streptomyces griseus aminopeptidase complexed with D-tryptophan | | Descriptor: | Aminopeptidase, CALCIUM ION, D-TRYPTOPHAN, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
2Y3R
 
 | | Structure of the tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P21 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-22 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
6OHY
 
 | | Chimpanzee SIV Env trimeric ectodomain. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallesen, J, Andrabi, R, de Val, N, Burton, D.R, Ward, A.B. | | Deposit date: | 2019-04-08 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Chimpanzee SIV Envelope Trimer: Structure and Deployment as an HIV Vaccine Template.
Cell Rep, 27, 2019
|
|
7KCR
 
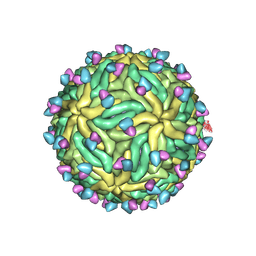 | | Cryo-EM structure of Zika virus in complex with E protein cross-linking human monoclonal antibody ADI30056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADI30056 Fab heavy chain variable region, ADI30056 Fab light chain variable region, ... | | Authors: | Sevvana, M, Rogers, T.F, Miller, A.S, Long, F, Klose, T, Beutler, N, Lai, Y.C, Parren, M, Walker, L.M, Buda, G, Burton, D.R, Rossmann, M.G, Kuhn, R.J. | | Deposit date: | 2020-10-07 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Zika Virus Specific Neutralization in Subsequent Flavivirus Infections.
Viruses, 12, 2020
|
|
2JAE
 
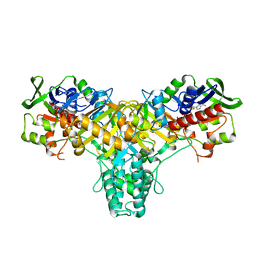 | | The structure of L-amino acid oxidase from Rhodococcus opacus in the unbound state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation.
J.Mol.Biol., 367, 2007
|
|
1TKJ
 
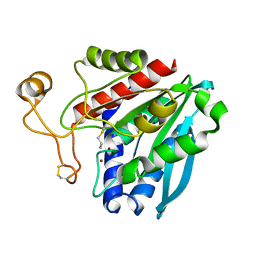 | | Streptomyces griseus aminopeptidase complexed with D-Methionine | | Descriptor: | Aminopeptidase, CALCIUM ION, D-METHIONINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
1ZK3
 
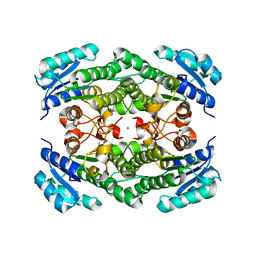 | | Triclinic crystal structure of the apo-form of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1TKH
 
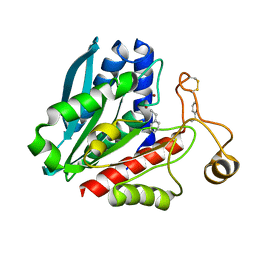 | | Streptomyces griseus aminopeptidase complexed with D-Phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, D-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
2JB3
 
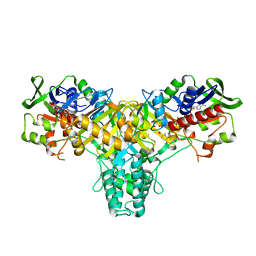 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with o-aminobenzoate | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
2EXI
 
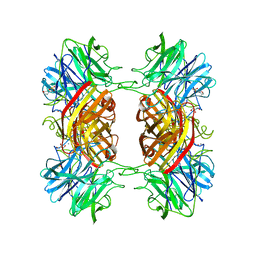 | | Structure of the family43 beta-Xylosidase D15G mutant from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
1XBU
 
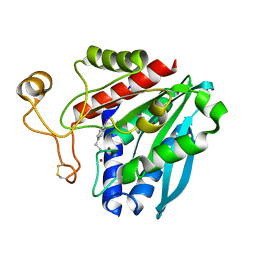 | | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, P-IODO-D-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine
To be Published
|
|
2JB2
 
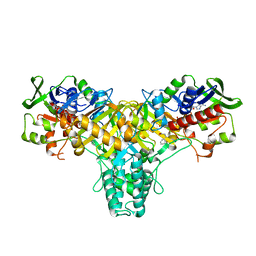 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with L-phenylalanine. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE, PHENYLALANINE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
2JB1
 
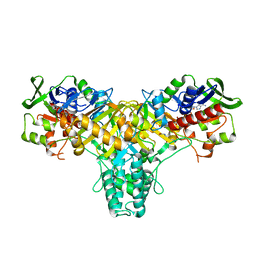 | | The L-amino acid oxidase from Rhodococcus opacus in complex with L- alanine | | Descriptor: | ALANINE, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
7JJN
 
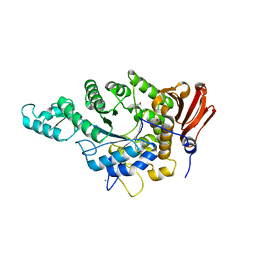 | | Eubacterium rectale Amy13B (EUR_01860) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Cerqueira, F.M. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structures of the GH13_36 amylases from Eubacterium rectale and Ruminococcus bromii reveal subsite architectures that favor maltose production
Amylase, 4, 2020
|
|
1MC9
 
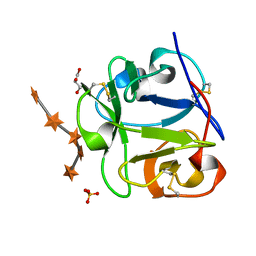 | | STREPROMYCES LIVIDANS XYLAN BINDING DOMAIN CBM13 IN COMPLEX WITH XYLOPENTAOSE | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, SULFATE ION, ... | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2002-08-06 | | Release date: | 2002-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1OD3
 
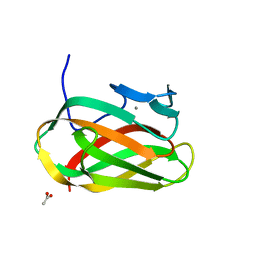 | | Structure of CSCBM6-3 From Clostridium stercorarium in complex with laminaribiose | | Descriptor: | ACETIC ACID, CALCIUM ION, PUTATIVE XYLANASE, ... | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
7LSU
 
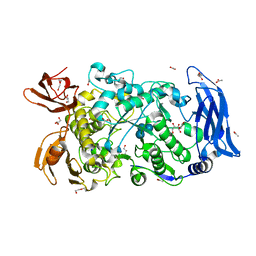 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LSA
 
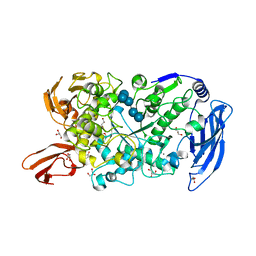 | | Ruminococcus bromii Amy12 with maltoheptaose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LST
 
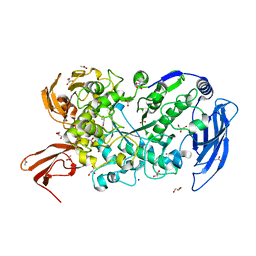 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LSR
 
 | | Ruminococcus bromii Amy12-D392A with maltoheptaose | | Descriptor: | CALCIUM ION, GLYCEROL, Pullulanase, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
1NAE
 
 | | Structure of CsCBM6-3 from Clostridium stercorarium in complex with xylotriose | | Descriptor: | CALCIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, putative xylanase | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G. | | Deposit date: | 2002-11-27 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains
J.Mol.Biol., 327, 2003
|
|
2EXJ
 
 | | Structure of the family43 beta-Xylosidase D128G mutant from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXK
 
 | | Structure of the family43 beta-Xylosidase E187G from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2NXY
 
 | | HIV-1 gp120 Envelope Glycoprotein(S334A) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
