4AUQ
 
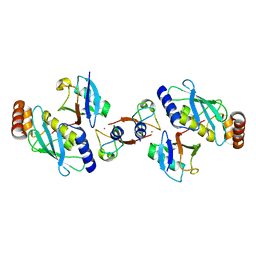 | | Structure of BIRC7-UbcH5b-Ub complex. | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 7, POLYUBIQUITIN-C, UBIQUITIN-CONJUGATING ENZYME E2 D2, ... | | Authors: | Dou, H, Buetow, L, Sibbet, G.J, Cameron, K, Huang, D.T. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.176 Å) | | Cite: | Birc7-E2 Ubiquitin Conjugate Structure Reveals the Mechanism of Ubiquitin Transfer by a Ring Dimer.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A49
 
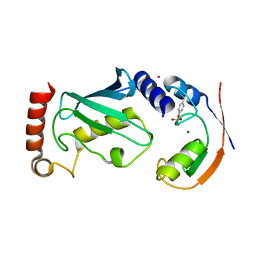 | | Structure of phosphoTyr371-c-Cbl-UbcH5B complex | | Descriptor: | E3 ubiquitin-protein ligase CBL, POTASSIUM ION, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-07 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl.
Nat. Struct. Mol. Biol., 19, 2012
|
|
4A4C
 
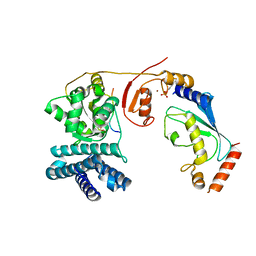 | | Structure of phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A4B
 
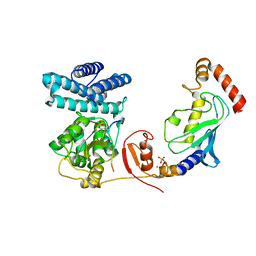 | | Structure of modified phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
2JF2
 
 | | Nucleotide substrate binding by UDP-N-acetylglucosamine acyltransferase | | Descriptor: | 1,2-ETHANEDIOL, ACYL-[ACYL-CARRIER-PROTEIN]--UDP-N-ACETYLGLUCOSAMINE O-ACYLTRANSFERASE, DI(HYDROXYETHYL)ETHER | | Authors: | Ulaganathan, V, Buetow, L, Hunter, W.N. | | Deposit date: | 2007-01-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nucleotide Substrate Recognition by Udp-N-Acetylglucosamine Acyltransferase (Lpxa) in the First Step of Lipid a Biosynthesis.
J.Mol.Biol., 369, 2007
|
|
2JF3
 
 | |
