2H4R
 
 | |
2DG9
 
 | |
2DUT
 
 | |
2AK4
 
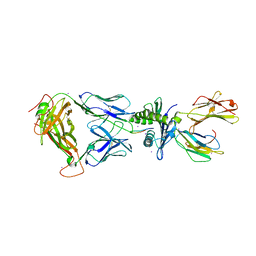 | | Crystal Structure of SB27 TCR in complex with HLA-B*3508-13mer peptide | | Descriptor: | Beta-2-microglobulin, EBV peptide LPEPLPQGQLTAY, HLA-B35 variant, ... | | Authors: | Tynan, F.E, Burrows, S.R, Buckle, A.M, Clements, C.S, Borg, N.A, Miles, J.J, Beddoe, T, Whisstock, J.C, Wilce, M.C, Silins, S.L, Burrows, J.M, Kjer-Nielsen, L, Konstenko, L, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T cell receptor recognition of a 'super-bulged' major histocompatibility complex class I-bound peptide
Nat.Immunol., 6, 2005
|
|
1B27
 
 | |
1B2U
 
 | |
1B3S
 
 | |
1B2S
 
 | |
2NW0
 
 | | Crystal structure of a lysin | | Descriptor: | ACETATE ION, PlyB | | Authors: | Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A Crystal Structure of the Catalytic Domain of PlyB, a Bacteriophage Lysin Active Against Bacillus anthracis.
J.Mol.Biol., 366, 2007
|
|
2PEE
 
 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Native State | | Descriptor: | GLYCEROL, SULFATE ION, Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
2PEF
 
 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Latent State | | Descriptor: | Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
7LZ6
 
 | | The Cryo-EM structure of a complex between GAD65 and b96.11 Fab | | Descriptor: | Glutamate decarboxylase 2, b96.11 Fab heavy chain, b96.11 Fab light chain | | Authors: | Reboul, C.F, Le, S.N, Williams, D.E, Buckle, A.M. | | Deposit date: | 2021-03-09 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure and dynamics of the autoantigen GAD65 in complex with the human autoimmune polyendocrine syndrome type 2-associated autoantibody b96.11
Biorxiv, 2024
|
|
1B21
 
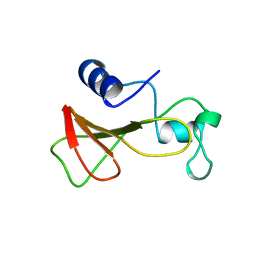 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B2X
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 7.5 FROM A CRYO_COOLED CRYSTAL AT 100K | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Harrison, P, Vaughan, C.K, Buckle, A.M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1BNJ
 
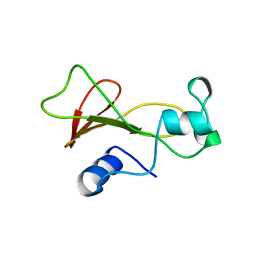 | | BARNASE WILDTYPE STRUCTURE AT PH 9.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
1B20
 
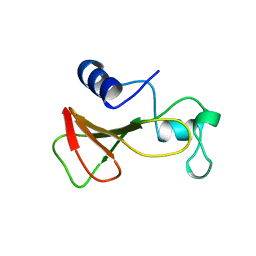 | | DELETION OF A BURIED SALT-BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B2Z
 
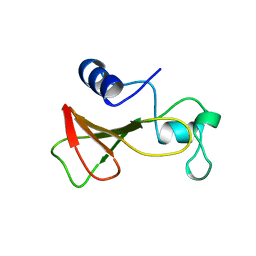 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1BNI
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 6.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
