2OCX
 
 | | Crystal structure of Se-Met fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-12-21 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
3ONE
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OND
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ONF
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with cordycepin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3'-DEOXYADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2HHC
 
 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
2HLH
 
 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | Nodulation fucosyltransferase, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
5D14
 
 | |
3SIX
 
 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase soaked with GDP-fucose | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, ... | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SIW
 
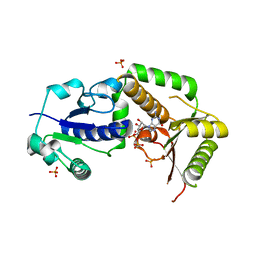 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase co-crystallized with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3P4J
 
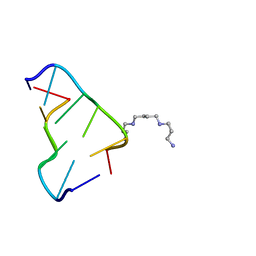 | | Ultra-high resolution structure of d(CGCGCG)2 Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE | | Authors: | Brzezinski, K, Brzuszkiewicz, A, Dauter, M, Kubicki, M, Jaskolski, M, Dauter, Z. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.55 Å) | | Cite: | High regularity of Z-DNA revealed by ultra high-resolution crystal structure at 0.55 A.
Nucleic Acids Res., 39, 2011
|
|
6TWT
 
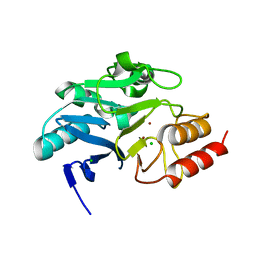 | | Crystal structure of N-terminally truncated NDM-1 metallo-beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Imiolczyk, B, Czyrko-Horczak, J, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
7O5M
 
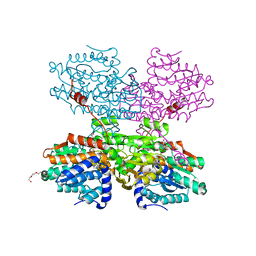 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Na+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7O5L
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3US6
 
 | | Crystal Structure of Histidine-containing Phosphotransfer Protein MtHPt1 from Medicago truncatula | | Descriptor: | Histidine-containing Phosphotransfer Protein type 1, MtHPt1 | | Authors: | Ruszkowski, M, Brzezinski, K, Jedrzejczak, R, Dauter, M, Dauter, Z, Sikorski, M, Jaskolski, M. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Medicago truncatula histidine-containing phosphotransfer protein: Structural and biochemical insights into the cytokinin transduction pathway in plants.
Febs J., 280, 2013
|
|
5TOV
 
 | |
5TOW
 
 | | Crystal structure of the inactive form of S-adenosyl-L-homocysteine hydrolase from Thermotoga maritima in ternary complex with NADH and Adenosine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE, ... | | Authors: | Czyrko, J, Brzezinski, K. | | Deposit date: | 2016-10-19 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | S-adenosyl-L-homocysteine hydrolase from a hyperthermophile (Thermotoga maritima) is expressed in Escherichia coli in inactive form - Biochemical and structural studies.
Int. J. Biol. Macromol., 104, 2017
|
|
4GS3
 
 | | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tencongensis solved ab initio | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Liebschner, D, Brzezinski, K, Dauter, M, Dauter, Z, Nowak, M, Kur, J, Olszewski, M. | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tengcongensis solved ab initio.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7QY6
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, wild type (WT EcAIII) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QYM
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-18 (R207V, D210P, S211W) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QYX
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-24 (R207A, D210S, S211T) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4Z1R
 
 | | Crystal structure of collagen-like peptide at 1.27 Angstrom resolution | | Descriptor: | Collagen-like peptide | | Authors: | Plonska-Brzezinska, M.E, Czyrko, J, Brus, D.M, Imierska, M, Brzezinski, K. | | Deposit date: | 2015-03-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Triple helical collagen-like peptide interactions with selected polyphenolic compounds.
Rsc Adv, 5, 2015
|
|
7ZD8
 
 | | Crystal structure of the R24E mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD7
 
 | | Crystal structure of the R24E/E352T double mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD9
 
 | | Crystal structure of the E352T mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD4
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase soaked with Cu+ ions | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
